A gene-centric comorbidity analysis for Epilepsy using results from SCAIView.
This repository contains code and resources used for the two analyses described in [1]
| [1] | Hoyt, C. T. and Domingo-Fernández, D., et al. (2018). A systematic approach for identifying shared mechanisms in epilepsy and its comorbidities. Database, 2018(June), bay050. |
First, a comorbidity analysis was performed with a literature-based approach based on overlapping co-occurrence of genes with Epilepsy and other diseases. Second, comorbidity analysis was performed on a mechanistic level with a comparative mechanistic enrichment approach based on a shared therapy, carbamazepime, between epilepsy and Alzheimer's disease.
- The resources and results folders contains all documents retrieved from SCAView queries as well as the tables and supplementary information presented in the paper
- Notebooks
- Epilepsy SCAIView Comorbidity Analysis outlines the calculations of pleiotropy rates using the resources in the repository. See below.
- Epidemiology vs. Literature-Based Comorbidity a comparison between epidemiological and literature/gene-based comorbidity analysis
- Epilepsy Corpora Overview gives a summary of the originally generate corpora compared to which articles were actually curated
- Epilepsy Knowledge Assembly Summary contains the summary of the different subgraphs used to generate Table 2 of the manuscript.
- Mechanism Enrichment Score Percentile Calculation describes the statistical calculations for chosen a threshold based on enrichment scores
- carbamazepine_targets.txt contains the gene list of targets of carbamazepine corresponding to Supplementary Text S1.
This section corresponds to the "Quantification of Gene Overlap" in the Methodology section of the paper. It uses screenshots as a guide to reproduce the analysis.
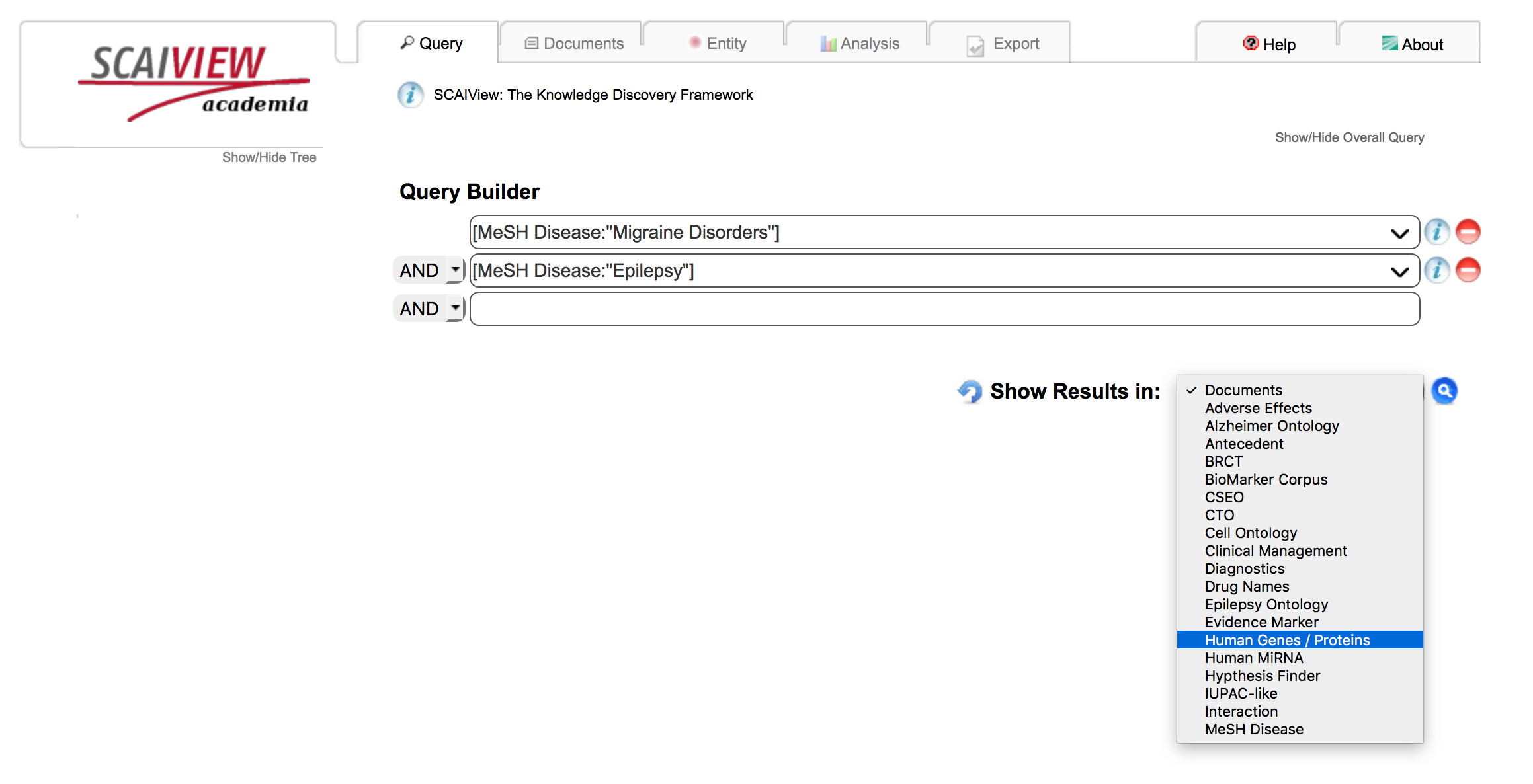
Build a query using SCAIView Query Interface the MeSH entry for Epilepsy and the target disease. In this example, "Migraine Disorders" is used. Select "Human Genes/Proteins" from the dropdown labeled "Show Results in:" in order to get the overlapping genes sorted by relevance (relative entropy).
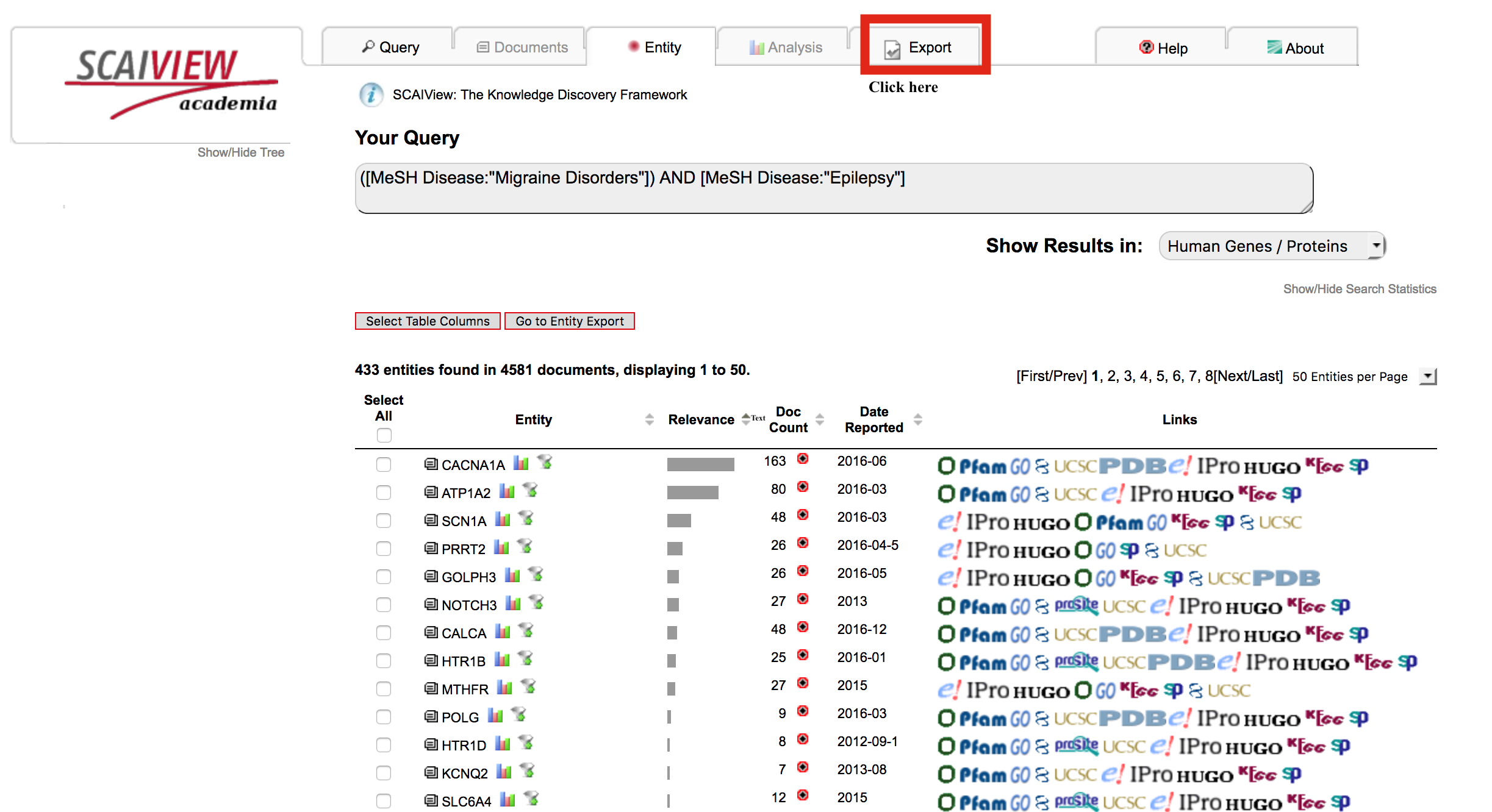
From the list of genes/proteins, the click the export tab to be given options on how to export the gene list and their associated information for programmatic use.
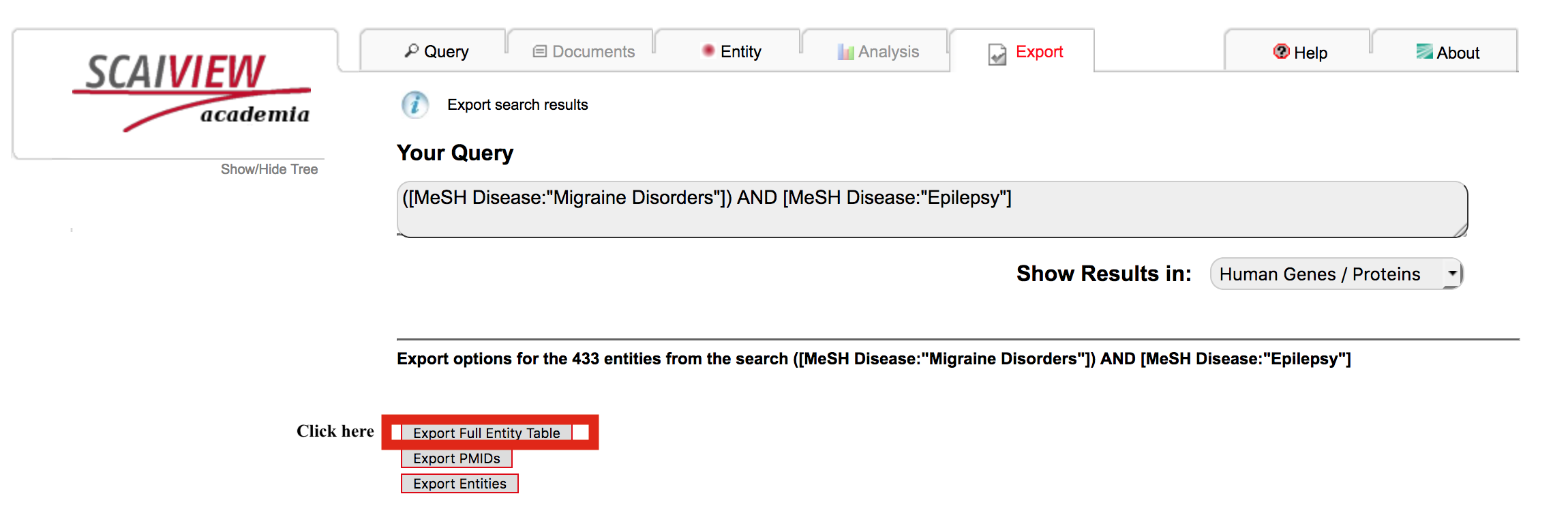
Click "Export Full Entity Table" in order to retrieve the genes/proteins, their relative entropies, their associated documents, and other useful information.
Repeat Steps 1-3 for several target diseases and stored in the results folder in this repository.
Clone this repository from GitHub with git clone https://github.com/neurommsig-epilepsy/EpiCom.git and cd into the directory. The Jupyter notebook included in this repostory, Epilepsy SCAIView Comorbidity Analysis, can be run from inside jupyter notebook in order to reproduce the analysis.
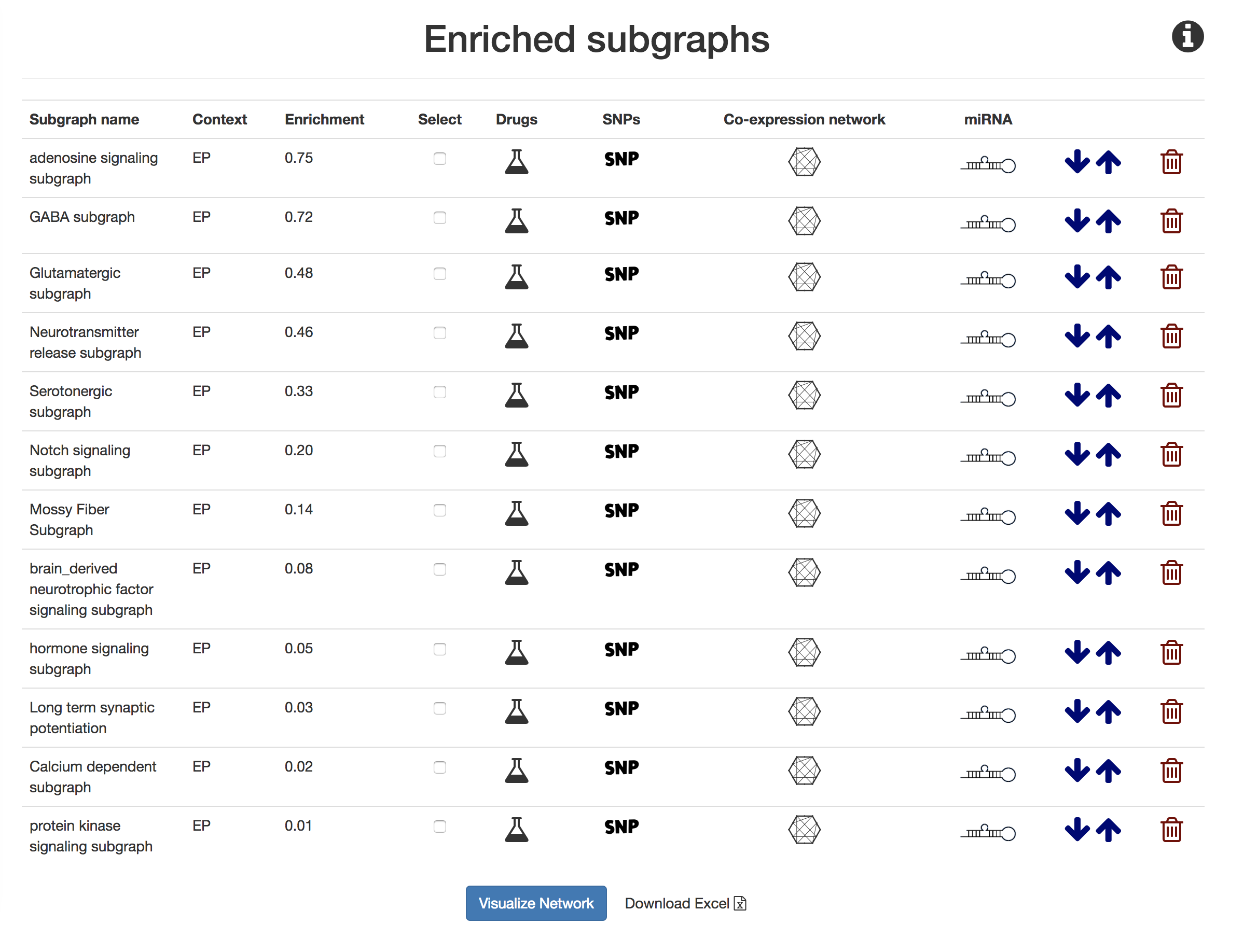
This section corresponds to the "Epilepsy Mechanism Enrichment" and "Comparative Mechanism Enrichment" headings in the Results section. It uses screenshots as a guide to reproduce the analysis.
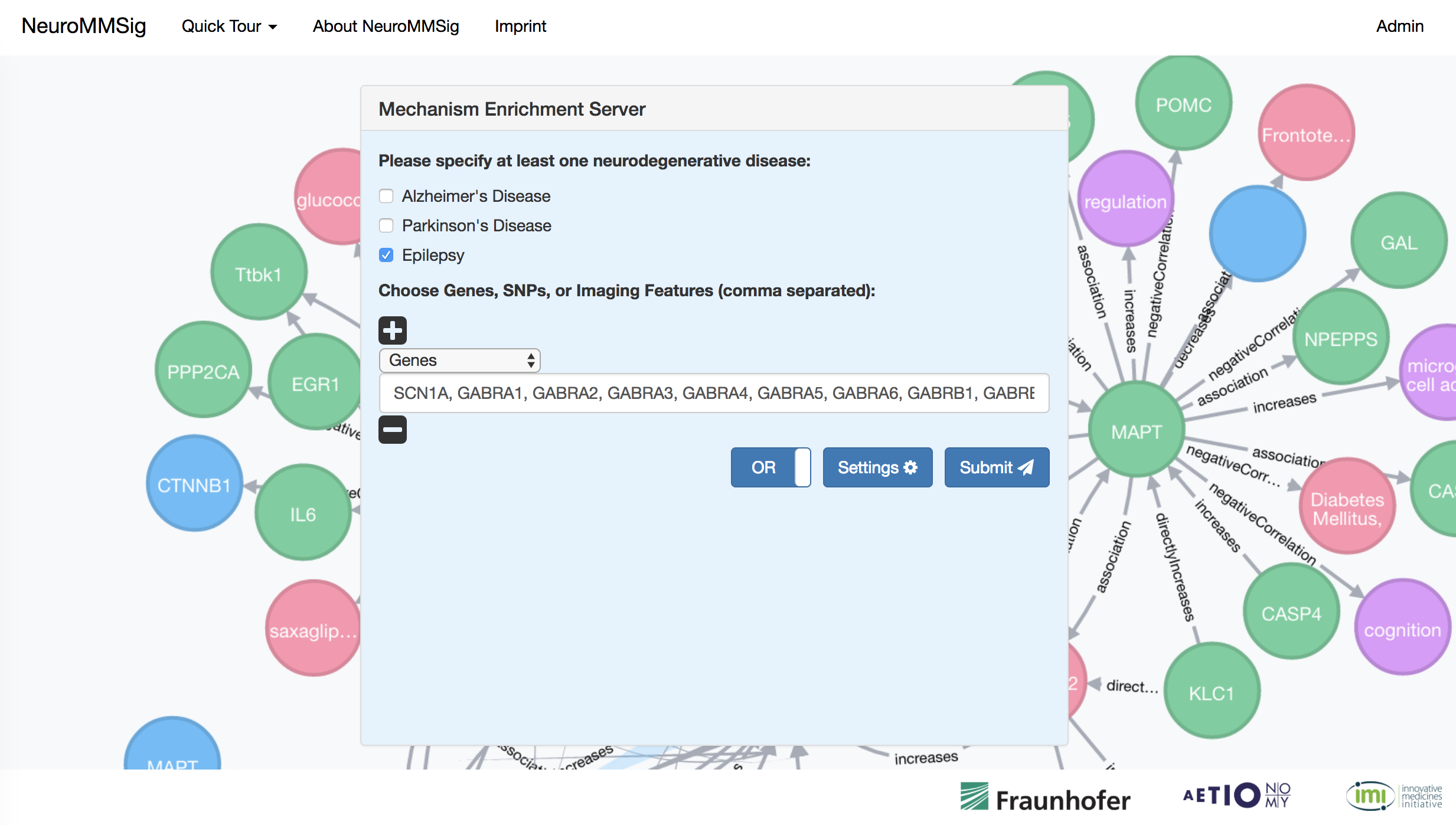
Submitted a query to NeuroMMSig with the protein targets of carbamazepime coming from PharmKGB against epilepsy.
Save the NeuroMMSig enrichment scores with the "Download Excel" button.
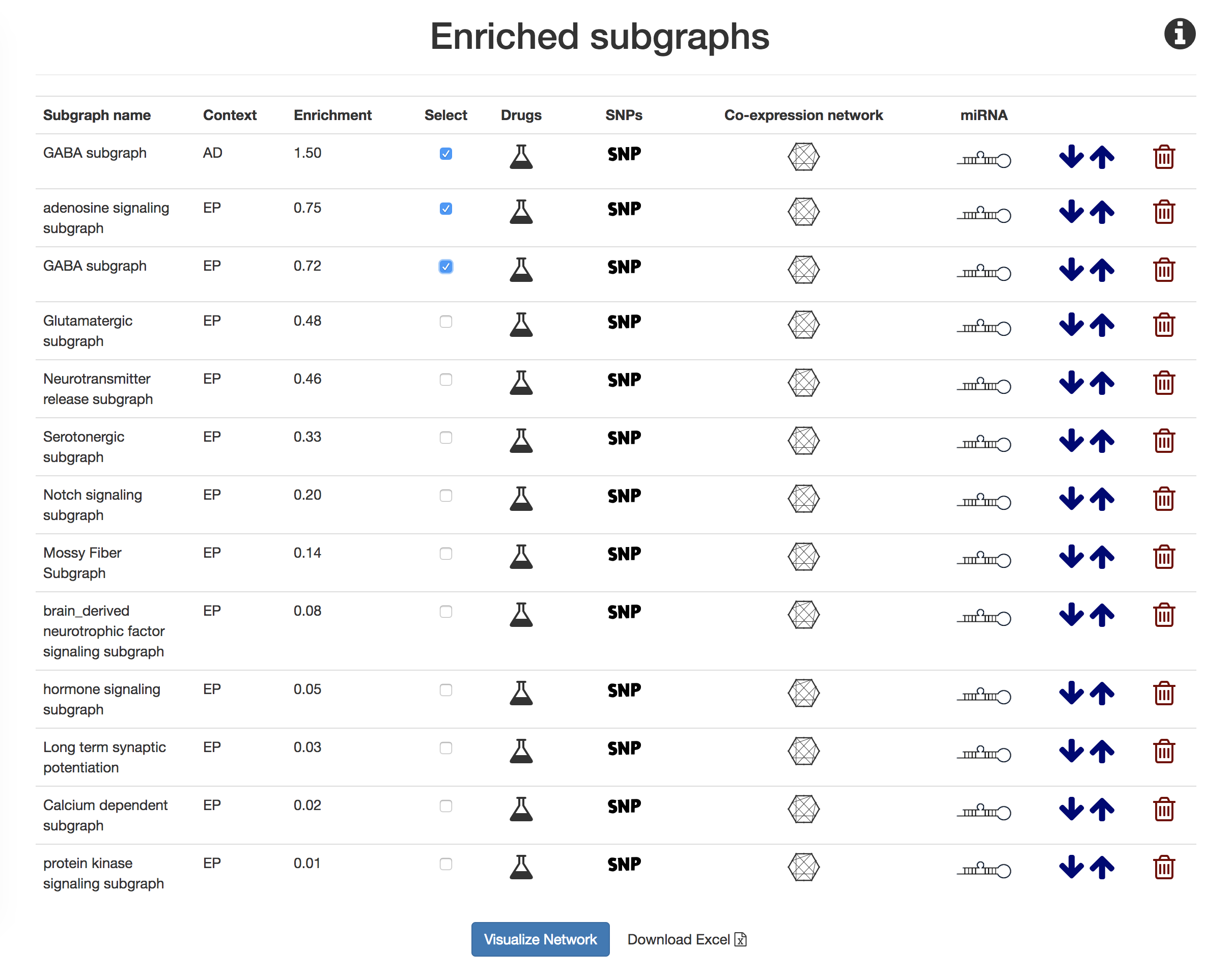
Repeat steps 1-2 for the Alzheimer's disease. Run the Jupyter notebook, Mechanism Enrichment Score Percentile Calculation, to identify an appropriate percentile cutoff for significant networks.
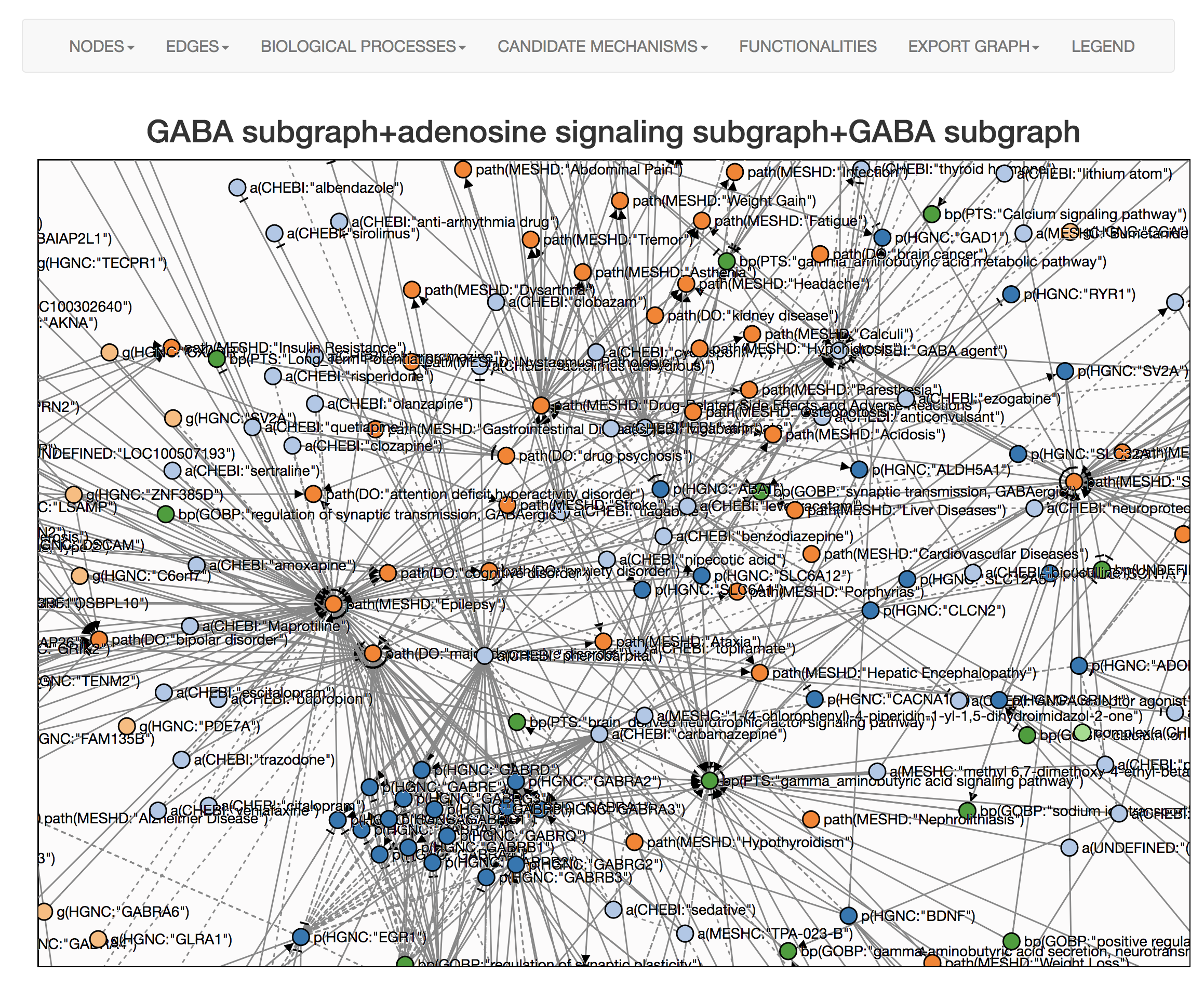
Perform enrichment with the combine context of Alzheimer's disease and epilepsy then choose all networks with enrichment scores above the cutoff.
Use the visualization to explore and generate hypotheses.