The technology of studying gene expression in space is gaining popularity. In particular, it is measured by adding different colors of fluorophore to the cell, which connects only to RNA produced from a fixed gene. Thus, by adding different colors of fluorophores to the cell, you can get an image with glowing dots. Each color corresponds to a gene, each point to a RNA molecule.
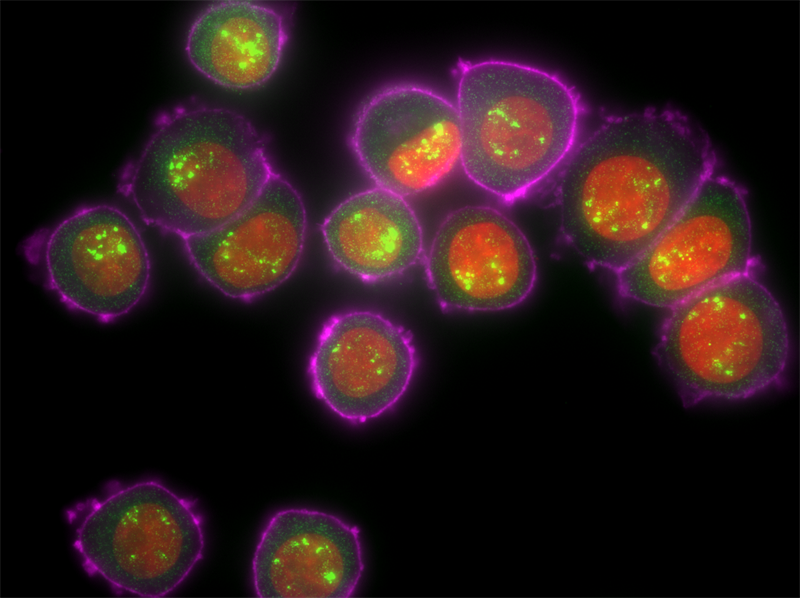
Image of simultaneous Stellaris RNA FISH and immunofluorescence assay.
HER2 mRNA molecules (green), protein marker HER2 (pink), DAPI (red) in
human SK-BR-3 cells (AV Orjalo Biosearch Tech.)
However, not all protocols can easily identify which cell which molecule belongs to which one. Thus, the formulation of the problem: to determine the boundaries of cells in the image from the data on the position of RNA molecules and genes from which these molecules were obtained. In addition, for the data under study there is information about the location of cell centers, obtained with the help of photographs of cell nuclei.
- Genes (represented as coordinates (x, y) in the plane)
- Example cell centers (cells consist of genes)
- Find the optimal cell boundaries in the picture
- Suppose that the cells have an oval shape, then you need to find the optimal size of the ellipse (i.e. its axis) and its center
- Then the cell can be represented as a set of points belonging to the same distribution. A two-dimensional normal distribution (it takes an average value and a spread of two coordinates, which is comparable to the center of the cell and its size) is good for this purpose
- We have to keep in mind that the data may be noisy. It will be expressed in the form of molecules, located far from the center of the cells. It can be described by a uniform distribution of
- We also assume that each cell has its own gene expression profile. It can be described by a multi-dimensional distribution.
- The whole task is to divide the points into clusters (cells). Some of them will be noise, others will be cells described by different distributions
- For this purpose, the Bayesian statistics methods were used in the project, in particular the EM algorithm
- Other signs are needed to describe the cells and the distribution that will describe them. They will be built into an EM algorithm already written
- We need additional algorithms to keep the cells from crossing
- We need to optimize the calculation algorithms