-
Notifications
You must be signed in to change notification settings - Fork 5
QuagmiR motif file
The motif-consensus.fa file will have the following structure:
>miRNA_name miRNA_motif
miRNA_consensus_sequence
>hsa-miR-21-5p TTATCAGACTGAT
TAGCTTATCAGACTGATGTTGA
where miRNA_motif will be the unique n-mer sequence that characterizes the uniquely the miRNA, and miRNA_consensus_sequence the canonical sequence of the miRNA we are comparing all isomiR to.
The following graph represents the percentage of unique motifs that can be generated from miRBase using different n-mer motif sequences in the center:
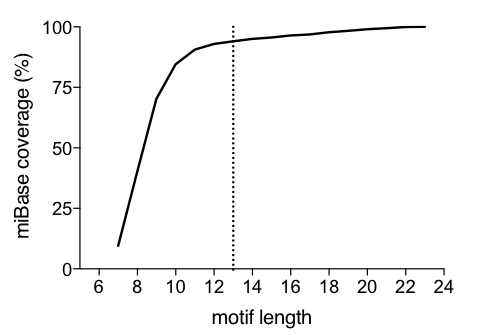
In order to increase sensitivity to RNA editing events that may impact the miRNA sequence, the miRNA_motif takes into account the inclusion of ambiguous nucleotides.
| Code | Bases |
|---|---|
| R | A or G |
| Y | C or T |
| S | G or C |
| W | A or T |
| K | G or T |
| M | A or C |
| B | C or G or T |
| D | A or G or T |
| H | A or C or T |
| V | A or C or G |
| N | any base |
To activate this option, change the option ambiguous_letters to True in the config.yaml file. Running QuagmiR on this mode increases its runtime. The activation of this mode also allows the matching of eventual sequence with ambiguous nucleotides from the fastq file.