- How to Get PHAT
- Working With PHAT Projects
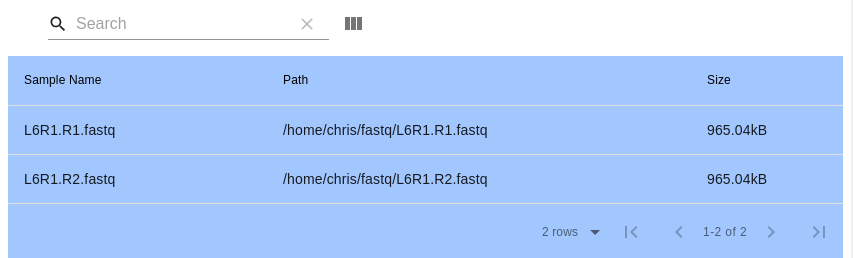
- Inputting Files
- Running Quality Control (QC) Reports
- Running Alignments
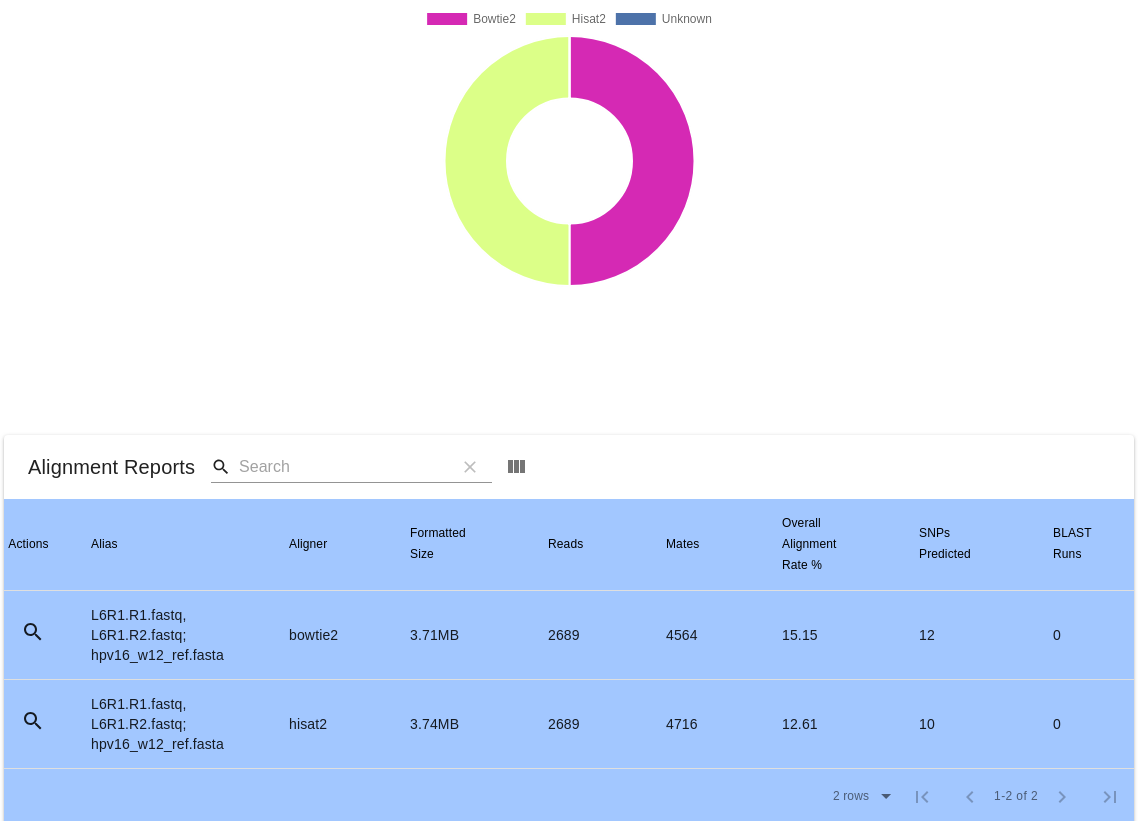
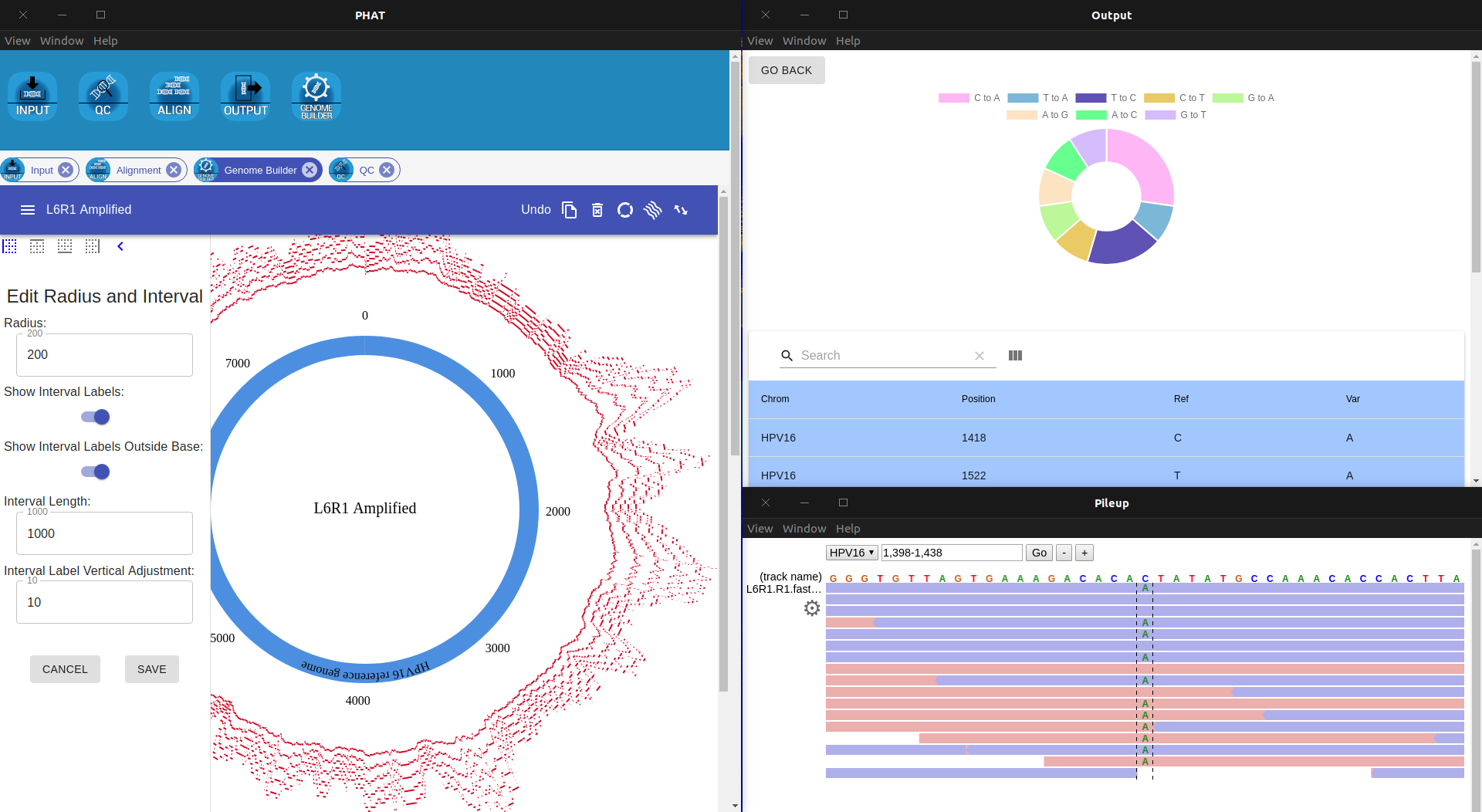
- Reports and Analyses
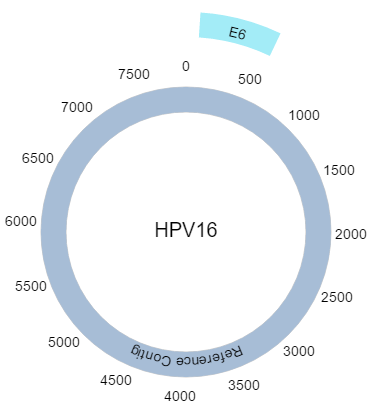
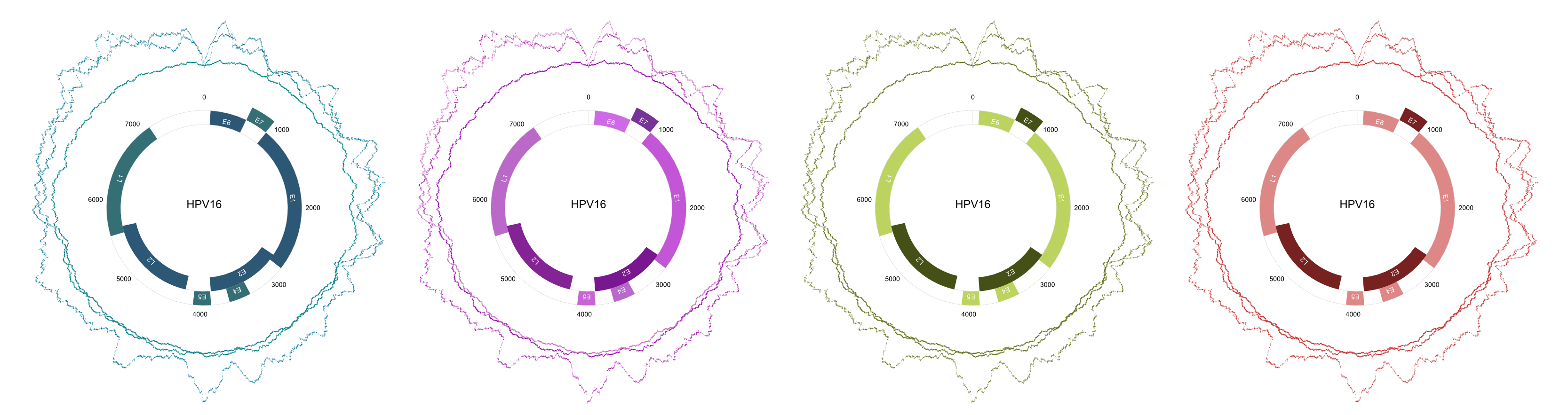
- Circular Visualization
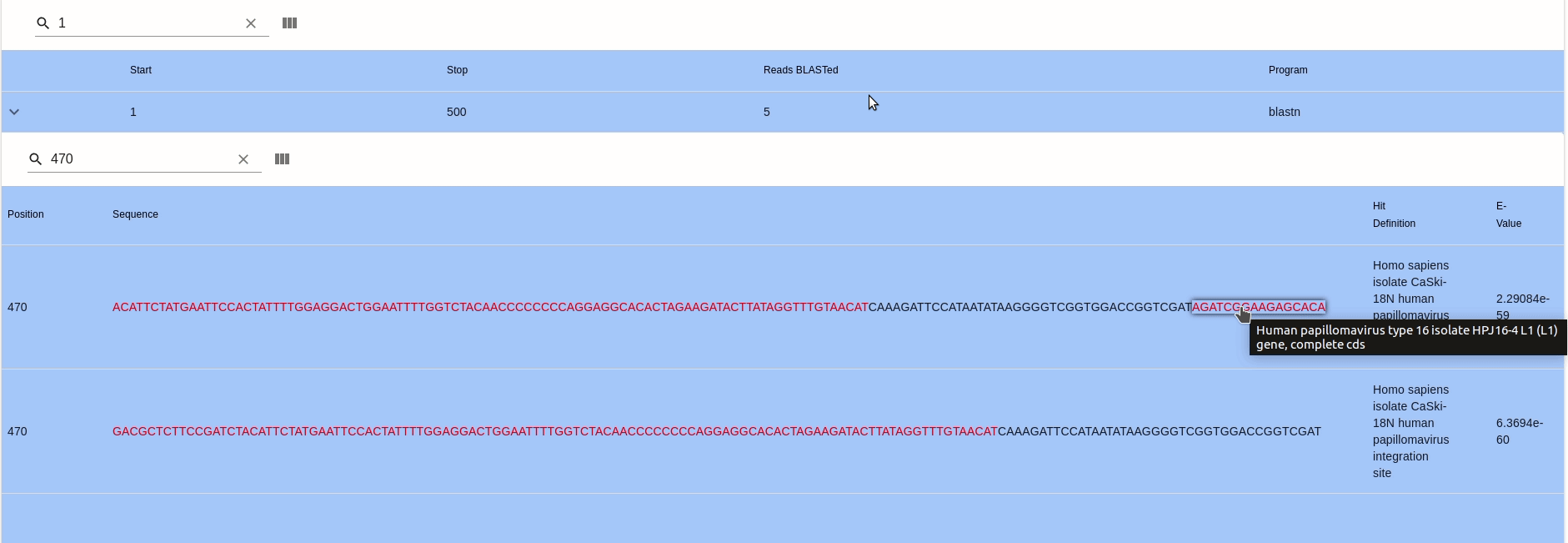
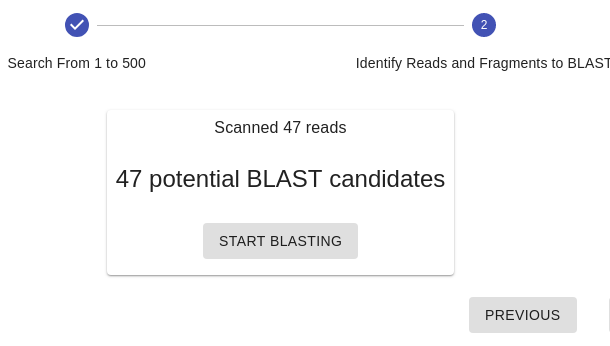
- BLASTing Alignments
The Pathogen-Host Analysis Tool (PHAT) is an application for processing and analyzing next-generation sequencing (NGS) data as it relates to relationships between pathogen and host organisms. PHAT provides quality control (QC) reporting on sequence files, alignment of sequence files against reference files, single-nucleotide polymorphism (SNP) prediction, linear and circular alignment viewing, and Excel and comma separated values (CSV) output.
PHAT is under development in the Zehbe Lab (https://www.zehbelab.com/) at the Thunder Bay Regional Health Research Institute (TBRHRI) and Lakehead University (LU) under the supervison of Dr. Ingeborg Zehbe. This work is supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant to Dr. Ingeborg Zehbe (#RGPIN-2015-03855) and a NSERC Alexander Graham Bell Canada Graduate Scholarship-Doctoral (CGS-D) to Robert Jackson (#454402-2014).
Project Supervisor/Principal Investigator: Dr. Ingeborg Zehbe, LU/TBRHRI Research Chair & Senior Scientist
Project Lead/Biologist: Robert Jackson, PhD Biotech Candidate
Project Lead/Software Developer: Chris Gibb, HBSc Comp Sci Student
Team Members: Mitchell Pynn, Shane Liu, Jeremy Braun, Nick Catanzaro, Zachary Moorman
How to Cite PHAT:
Gibb CM, Jackson R, Mohammed S, Fiaidhi J, Zehbe I. Pathogen-Host Analysis Tool (PHAT): an integrative platform to analyze next-generation sequencing data. Bioinformatics. 2018. DOI: 10.1093/bioinformatics/bty1003
| Build Statuses | master | beta | stable |
|---|---|---|---|
| Windows | |||
| Linux |