-
-
Notifications
You must be signed in to change notification settings - Fork 11.6k
Description
Proposed new feature or change:
(Scope) This suggestion is to modify "numpy.linalg.svd" and should give, for matrices where one axis is much larger than the other no less than a 20x speedup, and up to a 10,000x speedup.
If you have a matrix that is say 200 rows by 8 million columns, then when you multiply it by its transpose the result is 200x200 matrix. If you perform SVD on that you can determine both the left-singular vectors and the squares of the eigenvalues. It is symmetric and real, so according to this it is likely Hermitian.
(Versions) I am using numpy version 1.19.2, on python 3.8.12 in the following.
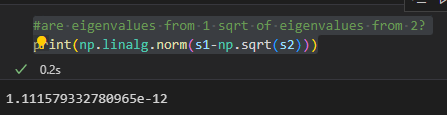
Here is example code to show the performance difference (using JuPyTeR):
import numpy as np
print(np.__version__)
---
A = np.random.rand(int(1e2), int(1e4))
AA = np.matmul(A,np.transpose(A))
---
%%timeit -n 3 -r 3
u1, s1, vt1 = np.linalg.svd(A)
---
u1, s1, vt1 = np.linalg.svd(A)
---
%%timeit -n 3 -r 3
u2, s2, vt2 = np.linalg.svd(AA)
s2b = np.sqrt(s2) #get eigens
rot_mat = np.matmul(u2, u1.T) #get rot for u2 --> u1
u2_in_u1 = np.matmul(rot_mat, u2) #perform rotation
B = np.matmul(np.diag(s2b),u2_in_u1) #get u1*s
B_inv = np.linalg.pinv(B) #find pinv
vt2b = np.matmul(B_inv,A) #isolate vt in u1 coords
the horizontal '---' indicate cell breaks
Here is a screenshot of the timing difference:
The image shows something like an order of 3-decade speedup for the operation. All the commented code in the SVD for AA, except for the square root, is extra. It maps V2 into V1. This is a strong over-estimate of how long it takes.
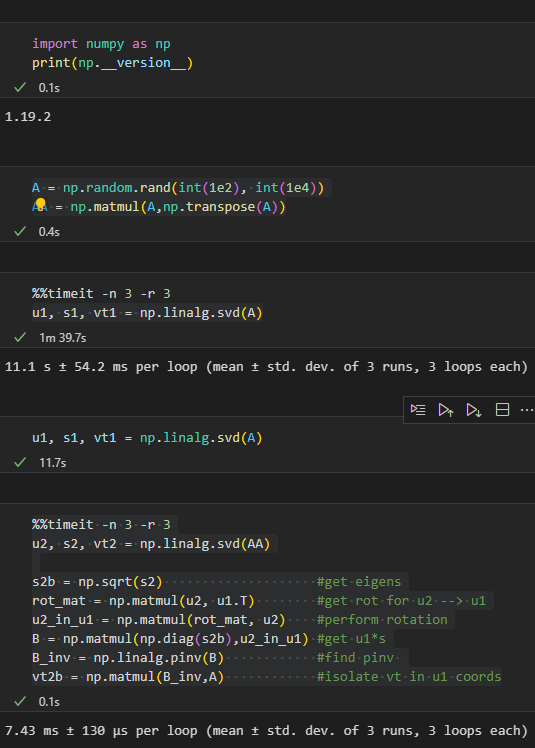
here is the code that shows the eigenvalues are compatible:
#are eigenvalues from 1 sqrt of eigenvalues from 2?
print(np.linalg.norm(s1-np.sqrt(s2)))
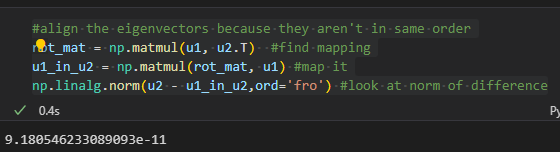
Here is the code that shows the left singular vectors are compatible:
#align the eigenvectors because they aren't in same order
rot_mat = np.matmul(u1, u2.T) #find mapping
u1_in_u2 = np.matmul(rot_mat, u1) #map it
np.linalg.norm(u2 - u1_in_u2,ord='fro') #look at norm of difference
Here is the image of the output
So what about the reduced dim options?
When I look for reduced dims, it gives this:
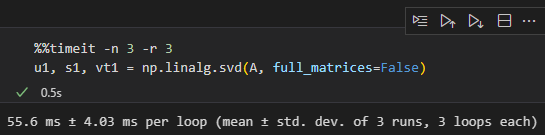
This takes 55.6/7.43 = 7.48 times longer to compute. I should be able to get the left singular matrix for the first decomposition, the rotation, using the results of the simpler matrix in less time.
If the matrix is scaled the other way, then some algebra and transposes should still yield similar speedups.
If the matrix is specified as Hermitian, (as symmetric and real should be) then the compute is done even more quickly.
I don't think every rectangular matrix multiplied by its transpose is Hermitian. This form of the real-valued A*A.T here can be contrived as a projection matrix, so it might be Hermitian.
(Disclaimer) I'm not sure if this is currently being done, but I think it isn't because of the decent multiple differences in timing (shown below). It has been some time since I took linear algebra, and so perhaps I am not as fresh as I could be there.