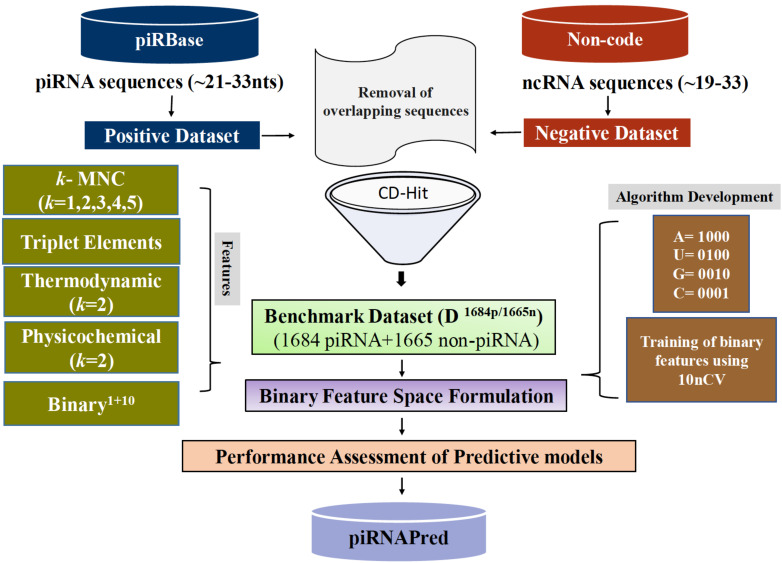
piRNAPred is an algorithm for the computational identification of piRNAs using features based on RNA sequence, structure, thermodynamic and physicochemical properties.
The PERL scripts, features used in the developemnt of piRNAPred algorithm along with the results and predictive models are provided here. Users can download individual files or can download the zipped version of piRNAPred (piRNAPred-GitHub.zip).
There are four folders within the piRNAPred algortihm folder:
-
Dataset files: FASTA and SVM format of main Dataset (D3349) and tissue specific datasets used in the development of piRNAPred.
-
models: SVM-based predictive models MDTTP-A+B+C, MDTTP-A+B+C+BINARY1+10 and models developed on the tissue specific datasets termed as testes-MDTTP-A+B+C+BINARY1+10 and ovary-MDTTP-A+B+C+BINARY1+10
-
PERL scripts: open-access to the PERL scripts used to calculate k-mer nucleotide composition (k-MNC), thermodynamic, SSTE and physicochemical properties in their respective sub-folders. Further, PERL scripts to call SVMlight and implement 10-fold cross-validation (10nCV).
-
Results: Performance of individual and hybrid features during 10nCV.
Monga, I., & Banerjee, I. (2019). Computational Identification of piRNAs Using Features Based on RNA Sequence, Structure, Thermodynamic and Physicochemical Properties. Current genomics, 20(7), 508–518. https://doi.org/10.2174/1389202920666191129112705