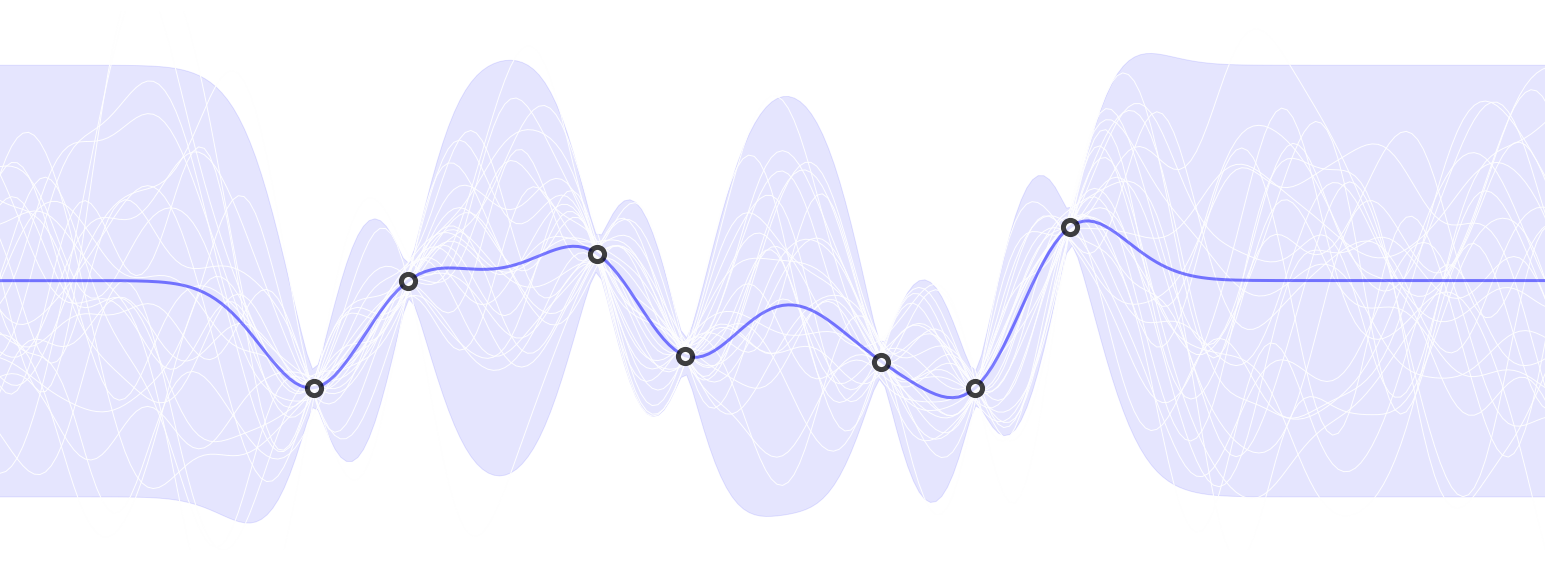
A gaussian process demo implemented by PyTorch, GPyTorch and NumPy.
A Gaussian process is a collection of random variables, any finite number of which have a joint Gaussian distribution.
Such a perspective comes from Rasmussen & Williams. Instead of the traditional linear regressions that produce numbers for prediction, a GP prediction is a distribution.
In standard linear regression, we have
where
A jointly Gaussian random variable
And here it comes the standard presentation of a Gaussian Process. Let's denote it as
As a well-known effective trick, the covariance function
The purpose of the Gaussian Process kernel function is to measure the similarity or relationship between data points in the input space. It defines how the outputs (function values) at different input points are correlated with each other. By doing so, it encodes our prior beliefs about the smoothness or structure of the underlying function we are trying to model.
To implement a Gaussian Process (GP) model, we first set up our kernel function, let's say the Radial Basis Function (RBF) kernel. We then collect training data and compute the kernel (covariance) matrix for the data points. The matrix will have dimensions n x n, where n is the number of training data points. Each element
The code gp_torch.py I present here is a more modern approach, simply using powerful PyTorch & GPyTorch. You can also check the detailed calculation in gp_calculate.py.