-
Notifications
You must be signed in to change notification settings - Fork 1.3k
Description
Describe the bug
I compared the orientation of a NIfTI file using MONAI, Nibabel and Mango. I found that MONAI's affine matrix does not match Nibabel's as well as Mango's. Since Mango provides correct result, let's just focus on Mango. In this case, I am pretty sure that MONAI is wrong with the (plus-minus) sign of first orientation (first row of affine matrix).
To Reproduce
To attach a NIfTI file here, I would like to put this NIfTI file on google drive, which I assure that it is safe to download. To open this NIfTI file with MONAI, I wrote the following (pseudo) code
from monai.transforms import LoadNifti
loader = LoadNifti(dtype=np.float32)
img, metadata = loader(<the path of this image.nii>)
print('img affine\n', metadata['affine'])
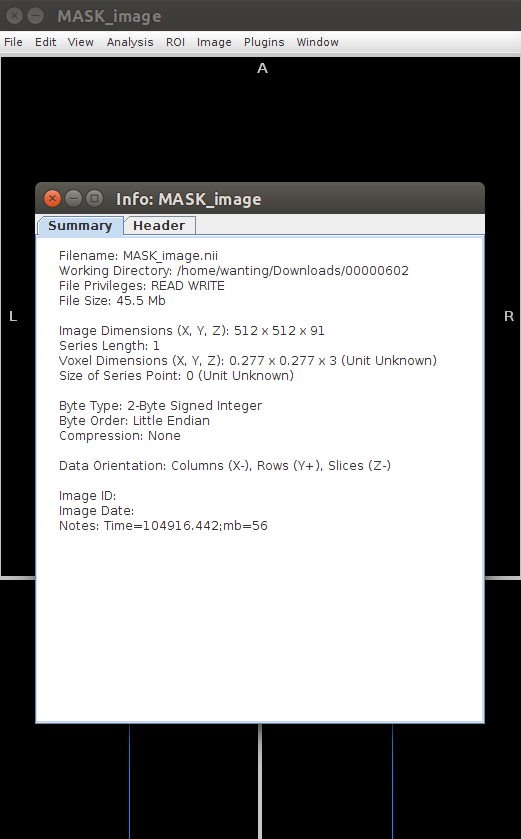
Then open this NIfTI file with Mango and use hot key: ctrl+i to see the summary info of nifti file.
Expected behavior
I expected the (plus-minus) sign of first three values of the diagonal of affine matrix from MONAI to be the same as data orientation in mango.
Screenshots
Using the above code, it will print the following result

Opening this nifti file, we will see such info (please notice the Data orientation in summary column.)
Environment (please complete the following information):
- OS: Ubuntu 18.04.3 LTS
- Python version: Python 3.6.9
- MONAI version [e.g. git commit hash]: 0.1.0
- CUDA/cuDNN version: 10.2
- GPU models and configuration: did not use GPU in this case
- Nibabel version: 3.0.0
Additional context
This nifti file is transformed from a DICOM file using dcm2niix.