-
Notifications
You must be signed in to change notification settings - Fork 12
Closed
Labels
Description
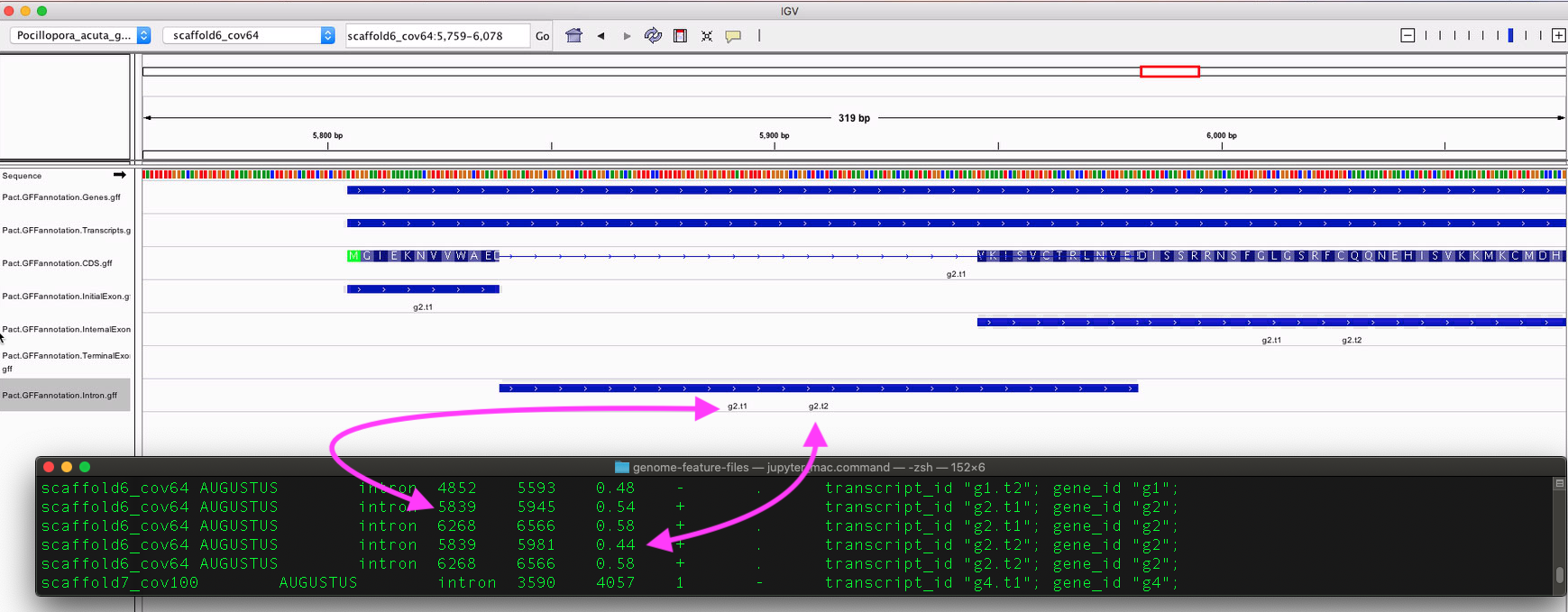
I visualized various P. acuta feature tracks in this IGV session (all features link to gannet so it should be viewable by others). Looking at the tracks, I noticed that introns were overlapping with CDS, which doesn't make any sense. I think this might be because IGV is taking introns from different transcripts that overlap with eachother and smashing them together.
If the introns, CDS, exons etc. for each transcript aren't mashed together, it would be easier to figure out if these tracks are valid. Is there something I need to do when generating tracks to ensure they're processed properly in IGV?