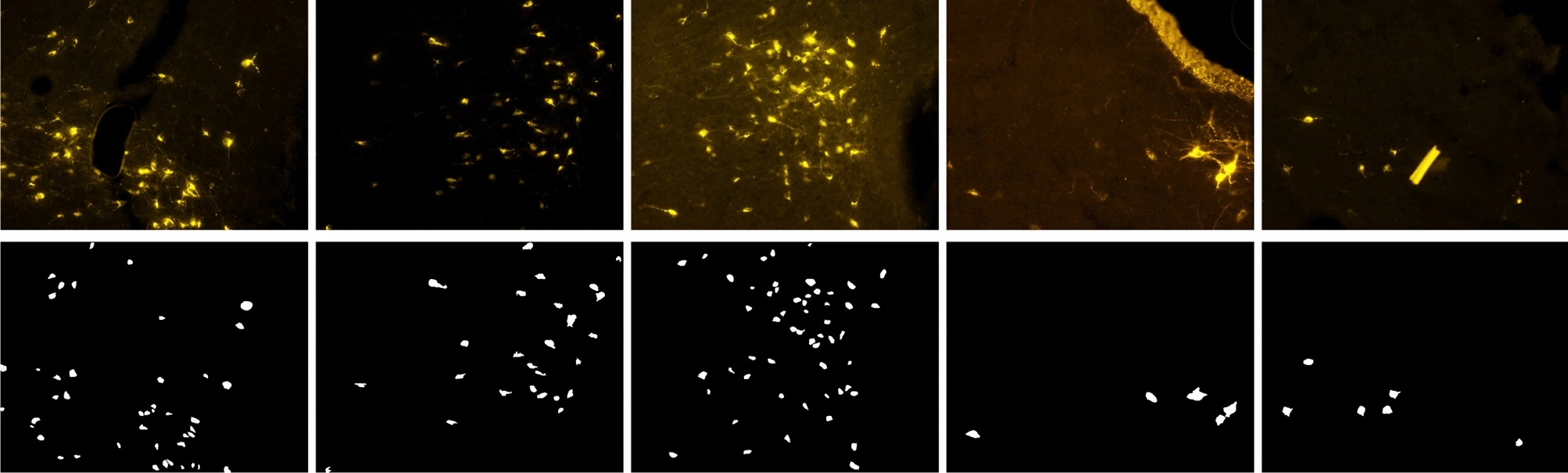
Image from original paper Automating cell counting in fluorescent microscopy through deep learning with c-ResUnet
Counting objects is a learning task common to many applications, from video sourveillance to agriculture 4.0, not to mention studies in life-sciences and medicine. However, this task is typically performed manually by domain experts, becoming very demanding in terms of time and human resources. Also, this increases the chances of errors due to distraction or fatigue.
This use-case deals with an approach to automate recognition and counting of objects in images. Specifically, we frame the problem as a semantic segmentation task and we use the c-Resunet network architecture, taking the Fluorescent Neuronal Cells dataset as a benchmark.
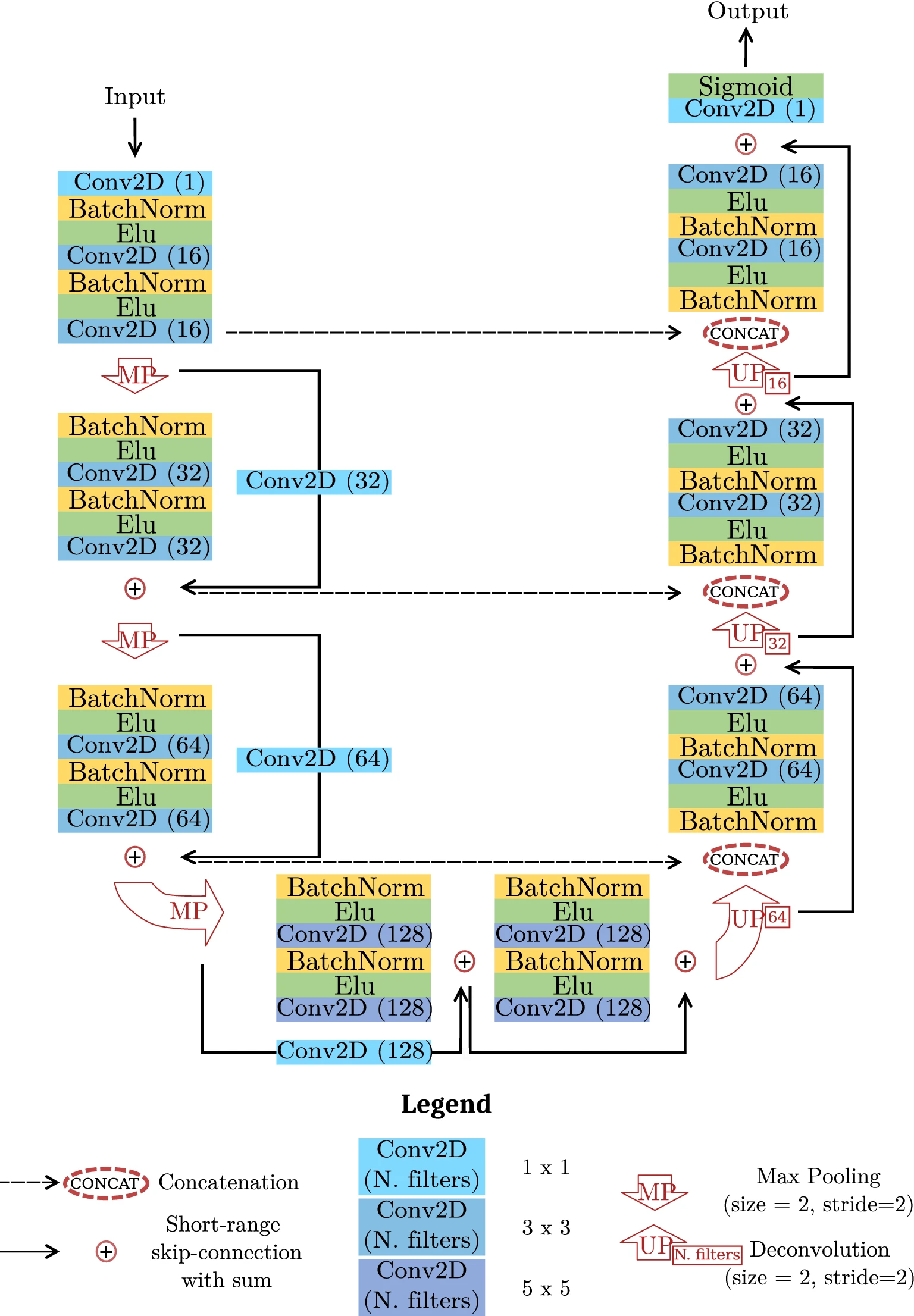
Image from original paper Automating cell counting in fluorescent microscopy through deep learning with c-ResUnet
The material is organized into notebooks that cover every stage of a realistic data analysis pipeline. In particular, a great deal of attention is devoted to EDA both to expose the challenges of the dataset and to evaluate the results. Likewise, technical aspects of the fastai implementation are detailed.
For more details on the implementations see blocks.py and utils.py scripts.
git clone git@github.com:clissa/object-counting-ML-INFN.git
After cloning the repository, simply follow the instructions in installation.txt to set up your workspace. Then download the data as described in the notebook 01. Exploratory Data Analysis.ipynb. Each step of the analysis is detailed in a dedicated notebook under the folder notebooks/.
Morelli, R., Clissa, L., Amici, R. et al. Automating cell counting in fluorescent microscopy through deep learning with c-ResUnet. Sci Rep 11, 22920 (2021).
Clissa, L. Supporting Scientific Research Through Machine and Deep Learning: Fluorescence Microscopy and Operational Intelligence Use Cases. PhD Thesis (2022)
Clissa, L. et al. Fluorescent Neuronal Cells. AMS Acta (2021)
Fluorescent Neuronal Cells dataset – part I, TDS Blog


