-
Notifications
You must be signed in to change notification settings - Fork 217
Description
hi,i meet a strange problem with running computematrix.
error:
Traceback (most recent call last):
File "/share/home/fanglab/liuyu/anaconda3/envs/chip/bin/computeMatrix", line 14, in
main(args)
File "/share/home/fanglab/liuyu/anaconda3/envs/chip/lib/python3.6/site-packages/deeptools/computeMatrix.py", line 421, in main
hm.computeMatrix(scores_file_list, args.regionsFileName, parameters, blackListFileName=args.blackListFileName, verbose=args.verbose, allArgs=args)
File "/share/home/fanglab/liuyu/anaconda3/envs/chip/lib/python3.6/site-packages/deeptools/heatmapper.py", line 264, in computeMatrix
verbose=verbose)
File "/share/home/fanglab/liuyu/anaconda3/envs/chip/lib/python3.6/site-packages/deeptools/mapReduce.py", line 85, in mapReduce
bed_interval_tree = GTF(bedFile, defaultGroup=defaultGroup, transcriptID=transcriptID, exonID=exonID, transcript_id_designator=transcript_id_designator, keepExons=keepExons)
File "/share/home/fanglab/liuyu/anaconda3/envs/chip/lib/python3.6/site-packages/deeptoolsintervals/parse.py", line 593, in init
self.parseGTF(fp, line)
File "/share/home/fanglab/liuyu/anaconda3/envs/chip/lib/python3.6/site-packages/deeptoolsintervals/parse.py", line 519, in parseGTF
if cols[2].lower() == self.transcriptID.lower():
IndexError: list index out of range
code:
computeMatrix scale-regions --metagene -p 20 -S result/rna_pe/BigWig/${rows}.bw
-R ~/genome/hg19/laji.gtf -b 0 -a 0 -m 10000
--missingDataAsZero --skipZeros -o mw/${rows}.mat.gz --sortRegions no --transcript_id_designator gene_id
~/genome/hg19/laji.gtf:
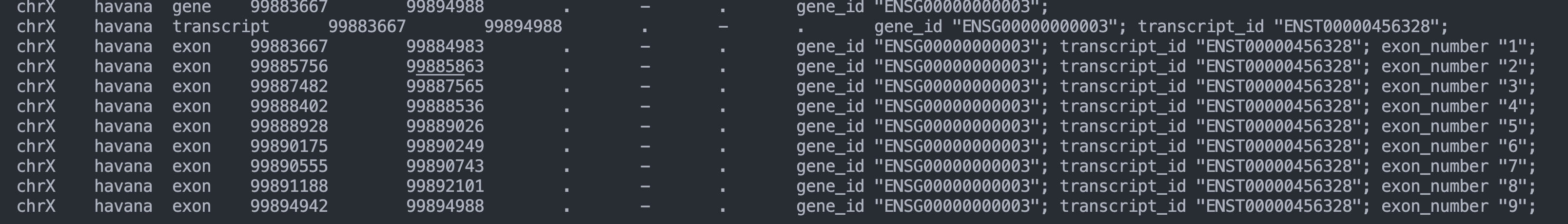
i compare my gtf with the gtf downloaded from gencode, i don't notice any difference.could you help me ? thanks alot!chrX havana gene 99883667 99894988 . - . gene_id "ENSG00000000003";
chrX havana transcript 99883667 99894988 . - . gene_id "ENSG00000000003"; transcript_id "ENST00000456328";
chrX havana exon 99883667 99884983 . - . gene_id "ENSG00000000003"; transcript_id "ENST00000456328"; exon_number "1";
chrX havana exon 99885756 99885863 . - . gene_id "ENSG00000000003"; transcript_id "ENST00000456328"; exon_number "2";
chrX havana exon 99887482 99887565 . - . gene_id "ENSG00000000003"; transcript_id "ENST00000456328"; exon_number "3";
chrX havana exon 99888402 99888536 . - . gene_id "ENSG00000000003"; transcript_id "ENST00000456328"; exon_number "4";
chrX havana exon 99888928 99889026 . - . gene_id "ENSG00000000003"; transcript_id "ENST00000456328"; exon_number "5";
chrX havana exon 99890175 99890249 . - . gene_id "ENSG00000000003"; transcript_id "ENST00000456328"; exon_number "6";
chrX havana exon 99890555 99890743 . - . gene_id "ENSG00000000003"; transcript_id "ENST00000456328"; exon_number "7";
chrX havana exon 99891188 99892101 . - . gene_id "ENSG00000000003"; transcript_id "ENST00000456328"; exon_number "8";
chrX havana exon 99894942 99894988 . - . gene_id "ENSG00000000003"; transcript_id "ENST00000456328"; exon_number "9";