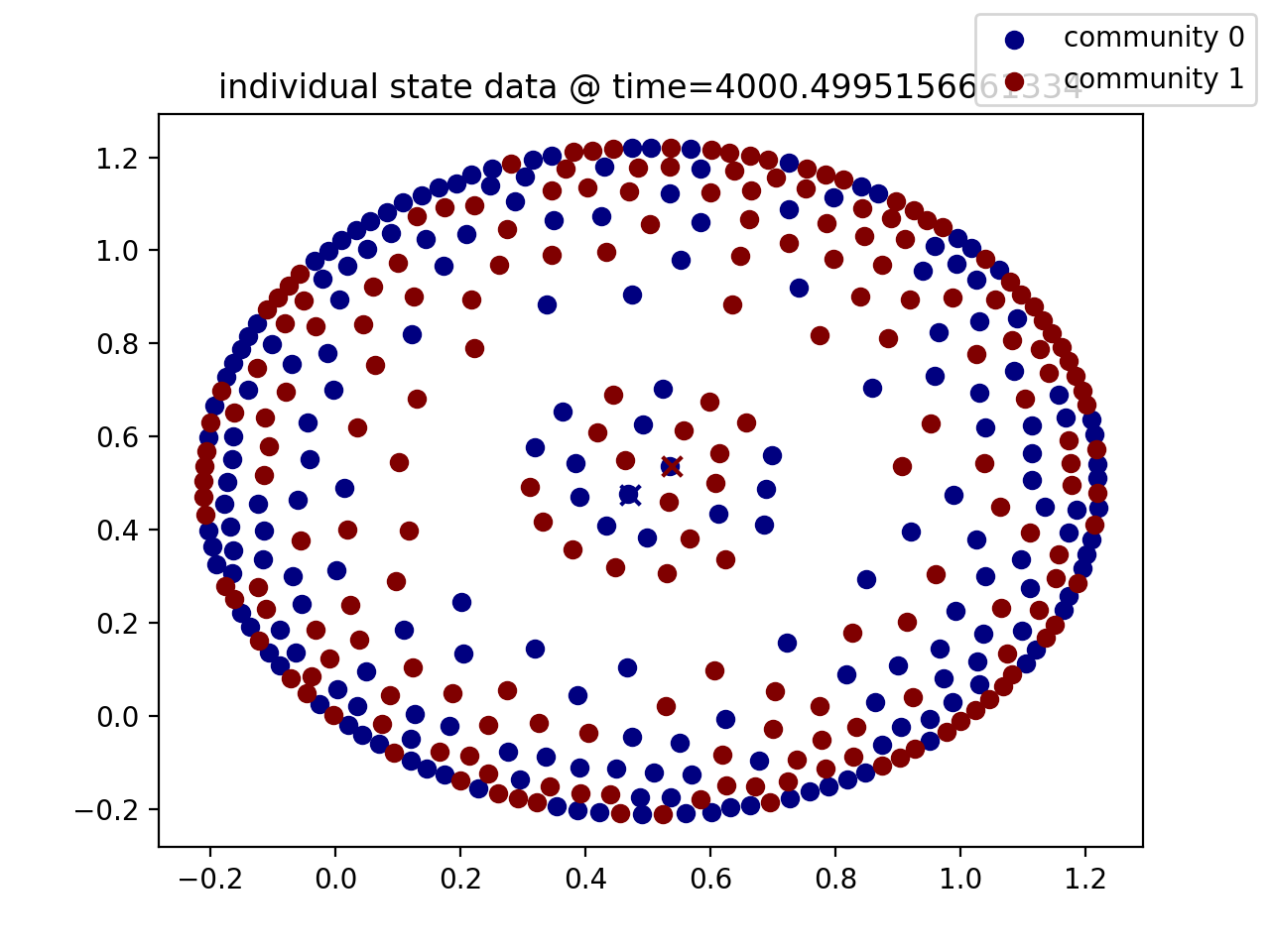
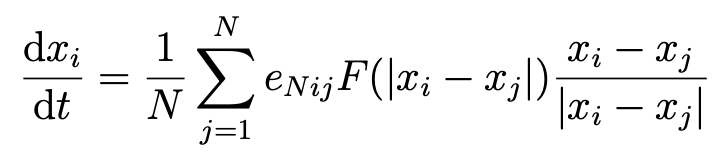aggregation_model is a package that provides a flexible numerical implementation of a particular aggregation model as explored in von Brecht et al.. This is an individual-based model that exhibits large-scale pattern formation, as in the below:
The system of differential equations used by this package is given below. The x_i variable represents the ith node's state data, N is the total number of nodes, e_{ij} is (1 or 0) the existence of an edge between node i and node j, and F is a pairwise interaction kernel. Typically, F(r) is assumed to be positive for small r and negative for large r; that is, individuals that are close tend to push each other away, and individuals that are far apart tend to draw each other in.
The main config file of this package is config.conf (which should be renamed to agg.conf by the user).
[SQL]
host = localhost
port = 3306
database = swarmsim
password = YOURPASS
The commands in the provided file init.sql should be run once; these schema create the swarmsim database and its associated tables:
- simulations, a parent table that contains fields for initial data from each simulation run;
- communities, a child table that stores community data;
- runs, a child table that stores per-run probabilities for each simulation group;
- statedata, another child table that stores the many-dimensional simulation state data;
- graphs, a child table for per-run graph data.
There are several components of this package:
- block_model.py, a module that offers a function for creating a stochastic block adjacency matrix and a class to properly wrap this matrix to feed into the other modules;
- odetools.py, a module that provides a function for generating initial conditions with added noise sampled from various random distributions and houses the logic for evolving the system of differential equations;
- plottools.py, a module that wraps the output of odetools and supplements it with methods that compute centers of mass from the data, plot state data by community, and call functions to record this data in a SQL database for future processing;
- sql_connector.py, an interface for mysql-connector-python that provides methods for easy reading of relevant data and writing of state data to a SQL database.
The module odetools.py can be run as a script to generate initial conditions and evolve the system of differential equations for a random graph:
if __name__ == '__main__':
C = [200, 200]
prob_array = np.array([[0.8, 0.2], [0.2, 0.8]])
grp = block_model.SBMGraph(C, prob_array)
init = generate_inits(grp, dims=2)
sol = odesolver(grp, init, final=4000, a=0, b=0.5)
This creates a random graph of 400 nodes with two "communities", each having 200 nodes of the total. This generates random initial conditions in two-dimensional space and then evolves the system of differential equations to time=4000. The output of odesolver is a plottools.SolutionWrapper instance, which comes with useful methods for analyzing and visualizing the resulting data:
M = compute_center()
computes each community's center of mass over time and assigns it to M. Additionally,
plot_state(time)
can be used to plot (by community) the particles' state data at time t. Note: if the system of equations is evolved in 1D, plot_state() returns a graph of particle data for all time t.
The module odetools.py contains a function run_simulation that uses many individual runs to span a grid of inner and outer probabilities. For example,
run_simulation(n_runs = 100, n_nodes = 100, a=0.5, b=0, inner_probs = [0.5,0.75,1], outer_probs = [0.6,0.7,0.8,0.9,1]
will cover the probability grid specified by inner_probs and outer_probs, where each probability pair has 100 individual runs from a network of 100 nodes (2 communities of 50 nodes each).
Due to being unable to hold this mass of data in memory all at once, it is expected the user has set up a SQL database from init.sql. The function automatically records simulation data in this structure. Incomplete past runs can be completed by using the resume=True flag; the most recent set of uncompleted runs are then finished.