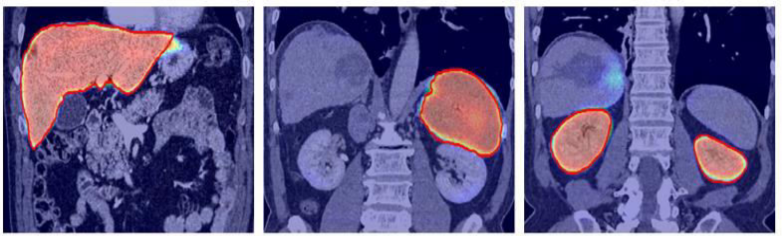
The objective of the project is to segment the organs and lables those organ in MRI Images.
Organ-Segmentation-and-Labeling-in-MRI-Images
Due to increase in a large dataset of human body MRI, it is tedious and time-consuming to segment each organ manually and then automatically analyze patient-specific organ. To make the process digitized computerized analysis of these images requires accurate segmentation of anatomical regions. The segmented images contain homogeneous, non-overlapping, semantically meaningful regions of the similar attribute. Classification is used to label each segmented organ. Intensity-based image segmentation is feasible when there is a large difference between the intensities of the object of interest and its background.
It is not an easy task to automatically segment MRI because MRI is imperfect or corrupted by noise and other image artifacts. Segmentation results are affected by:
Inhomogeneous artifacts: that cause shading when simple gray level based segmentation is used.
Partial volume effect: when a single pixel is covered by multiple tissues then different object boundaries become blurred.
- Step 1: Data collection - In this phase different organ MRIs will be collected and labeled. It involves the collection of MRI of Kidney, Lung, Liver, Gallbladder, Pancreas, Spleen, Stomach
- Step 2: Data Pre-processing - In this phase, all images will be processed for removing noise, intensity normalization, bias-field correction.
- Step 3: Constructing model -In this phase, some pre-trained models like ResNet will be explored on MRI images and creation of own CNN models.
- Step 4: Training and Experimentation -In this stage , the pre-processed images will be feed as input to the network. After training, the network will be tested on other MRI images for segmentation
- Step 1: The MRI photos are collected from url: https://www.mr-tip.com/serv1.php?type=db1&dbs=Abdominal%20Imaging
- Step 2: CNN is trained using python keras and tensor flow to create a weights files from training dataset.
- Step 3: Testing image is given to trained CNN and checked which class it is predicting the image to be.
Measures such as Precision, Recall, True Positive rate, root mean square surface rate can be used.
Anaconda with spyder is used for CNN which uses python libraries of keras and tensorflow. The hardware needed will be of multi core fast processor or a GPU machine to train on large dataset with epochs more than 40. This will take training time nearly equal to 1 hour. After saving these weights we get a trained model and this is used to predict new image class. The CNN can be multi layer with 3-4 hidden layers and 3 classes or categories with Relu (Rectified Linear Unit) activation function. The loss function used will be adams optimizer and categorial cross entropy.
Training will be conducted on NVIDIA GPU
Can be implemeted in Medical Sector which can save lots of time of doctor of visulization of MRI and can make system more efficient.
In case of any discussion or any suggestion reach us @
- mail- sau29gupta@gmail.com
After successfully implementation of the project we will deploy it and too have a plan to deploy it on a separate webpage.
We are open to contributions. If you wanna contribute you must have a knowldege of CNN and basic EDA part of Machine Learning and your suggestions are welcomed heartly.
Saurabh Kumar
Will be released under Apache Community LICENSE.
We are under the working, up and running state of the project hope we will come out with facinating result soon.