-
Notifications
You must be signed in to change notification settings - Fork 261
Description
Description
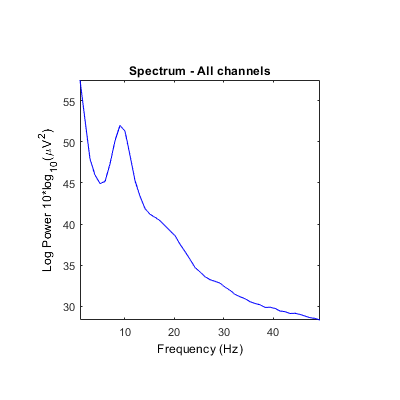
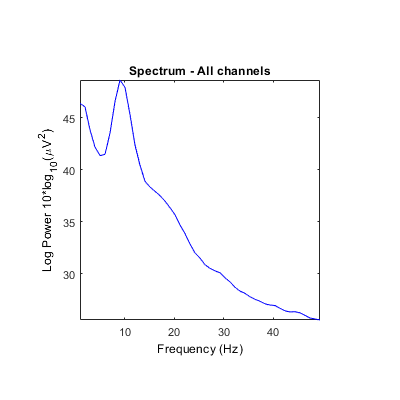
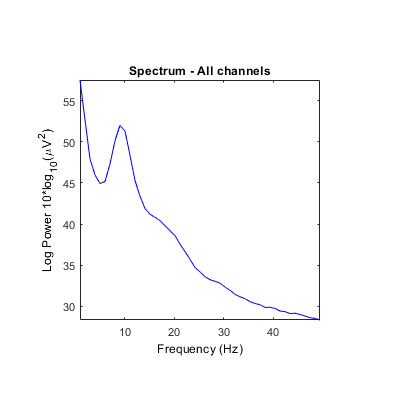
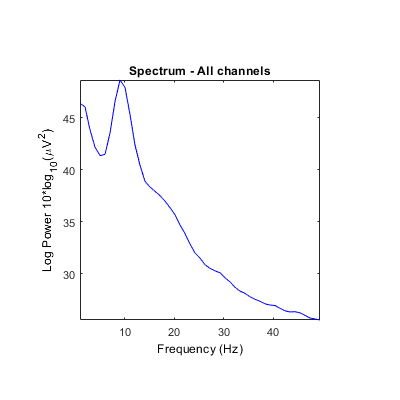
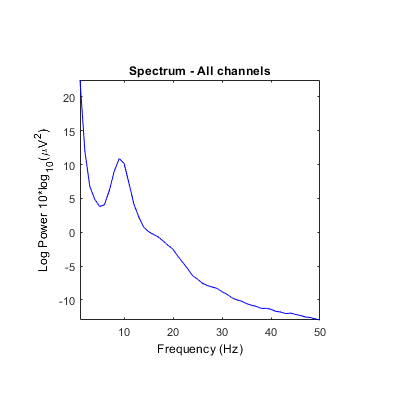
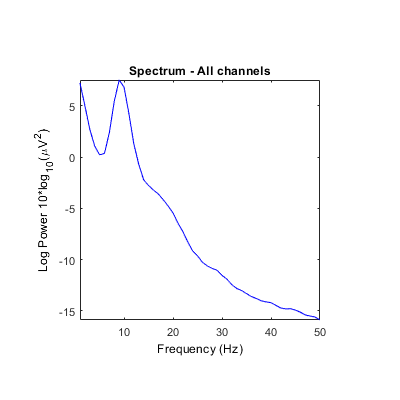
Unclear what scales/units are returned by some STUDY power spectrum modes. Is it either absolute power or dB? Units in ylabel don't seem to correspond (see images below). It always indicates dB even though values and settings are different.
And outputs should potentially be checked to make sure the correct values are returned. There are a lot of 'if' conditions, conversions, and methods employed, so it's hard to keep track (e.g., psd computed in dB by spectcomp in spectopo with either pwelch or spec method depending on presence/absence of signal processing toolbox, then converted to dB in spectopo, and converted again to abs power in std_spec).
Also, calculating psd is extremly long compared to when I compute it with pwelch directly in Matlab. Not sure why.
Also, for FFT, there doesn't seem to be a difference if you chose the option 'output' 'power' or not.
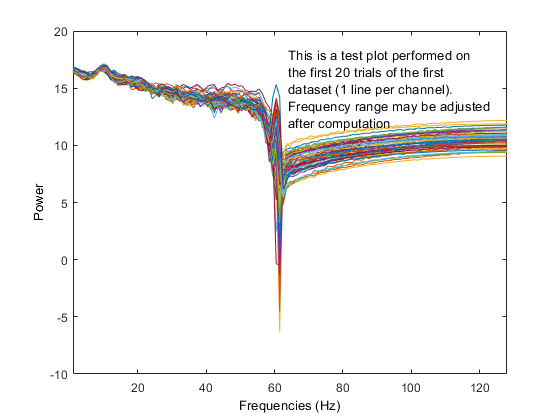
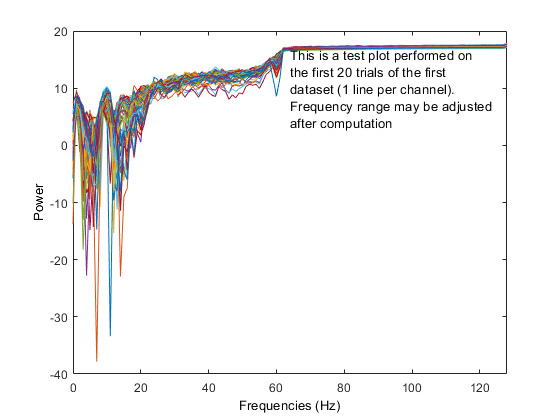
Using the 'test' feature in the precompute window, different plots are generated when using the log 'on' option (compared to above after precomputation).
#### Steps to Reproduce
- Load a study
- precompute channel power spectra:
[STUDY, ALLEEG] = std_precomp(STUDY, ALLEEG, {},'savetrials','on','recompute','on','spec','on','specparams',...
{'specmode','fft','logtrials','off'});
STUDY = pop_specparams(STUDY, 'averagechan','on', 'freqrange',[1 50]);
STUDY = std_specplot(STUDY,ALLEEG,'channels',{'Fp1','AF7','AF3','F1','F3','F5','F7','FT7','FC5','FC3','FC1','C1','C3','C5','T7','TP7','CP5','CP3','CP1','P1','P3','P5','P7','P9','PO7','PO3','O1','Iz','Oz','POz','Pz','CPz','Fpz','Fp2','AF8','AF4','AFz','Fz','F2','F4','F6','F8','FT8','FC6','FC4','FC2','FCz','Cz','C2','C4','C6','T8','TP8','CP6','CP4','CP2','P2','P4','P6','P8','P10','PO8','PO4','O2'}, 'design', 1);
[STUDY, ALLEEG] = std_precomp(STUDY, ALLEEG, {},'savetrials','on','recompute','on','spec','on','specparams',...
{'specmode','fft','logtrials','on'});
STUDY = pop_specparams(STUDY, 'averagechan','on', 'freqrange',[1 50]);
STUDY = std_specplot(STUDY,ALLEEG,'channels',{'Fp1','AF7','AF3','F1','F3','F5','F7','FT7','FC5','FC3','FC1','C1','C3','C5','T7','TP7','CP5','CP3','CP1','P1','P3','P5','P7','P9','PO7','PO3','O1','Iz','Oz','POz','Pz','CPz','Fpz','Fp2','AF8','AF4','AFz','Fz','F2','F4','F6','F8','FT8','FC6','FC4','FC2','FCz','Cz','C2','C4','C6','T8','TP8','CP6','CP4','CP2','P2','P4','P6','P8','P10','PO8','PO4','O2'}, 'design', 1);
[STUDY, ALLEEG] = std_precomp(STUDY, ALLEEG, {},'savetrials','on','recompute','on','spec','on','specparams',...
{'specmode','fft','logtrials','off', 'output', 'power'});
STUDY = pop_specparams(STUDY, 'averagechan','on', 'freqrange',[1 50]);
STUDY = std_specplot(STUDY,ALLEEG,'channels',{'Fp1','AF7','AF3','F1','F3','F5','F7','FT7','FC5','FC3','FC1','C1','C3','C5','T7','TP7','CP5','CP3','CP1','P1','P3','P5','P7','P9','PO7','PO3','O1','Iz','Oz','POz','Pz','CPz','Fpz','Fp2','AF8','AF4','AFz','Fz','F2','F4','F6','F8','FT8','FC6','FC4','FC2','FCz','Cz','C2','C4','C6','T8','TP8','CP6','CP4','CP2','P2','P4','P6','P8','P10','PO8','PO4','O2'}, 'design', 1);
[STUDY, ALLEEG] = std_precomp(STUDY, ALLEEG, {},'savetrials','on','recompute','on','spec','on','specparams',...
{'specmode','fft','logtrials','on', 'output', 'power'});
STUDY = pop_specparams(STUDY, 'averagechan','on', 'freqrange',[1 50]);
STUDY = std_specplot(STUDY,ALLEEG,'channels',{'Fp1','AF7','AF3','F1','F3','F5','F7','FT7','FC5','FC3','FC1','C1','C3','C5','T7','TP7','CP5','CP3','CP1','P1','P3','P5','P7','P9','PO7','PO3','O1','Iz','Oz','POz','Pz','CPz','Fpz','Fp2','AF8','AF4','AFz','Fz','F2','F4','F6','F8','FT8','FC6','FC4','FC2','FCz','Cz','C2','C4','C6','T8','TP8','CP6','CP4','CP2','P2','P4','P6','P8','P10','PO8','PO4','O2'}, 'design', 1);
[STUDY, ALLEEG] = std_precomp(STUDY, ALLEEG, {},'savetrials','on','recompute','on','spec','on','specparams',...
{'specmode','psd','logtrials','off'});
STUDY = pop_specparams(STUDY, 'averagechan','on', 'freqrange',[1 50]);
STUDY = std_specplot(STUDY,ALLEEG,'channels',{'Fp1','AF7','AF3','F1','F3','F5','F7','FT7','FC5','FC3','FC1','C1','C3','C5','T7','TP7','CP5','CP3','CP1','P1','P3','P5','P7','P9','PO7','PO3','O1','Iz','Oz','POz','Pz','CPz','Fpz','Fp2','AF8','AF4','AFz','Fz','F2','F4','F6','F8','FT8','FC6','FC4','FC2','FCz','Cz','C2','C4','C6','T8','TP8','CP6','CP4','CP2','P2','P4','P6','P8','P10','PO8','PO4','O2'}, 'design', 1);
[STUDY, ALLEEG] = std_precomp(STUDY, ALLEEG, {},'savetrials','on','recompute','on','spec','on','specparams',...
{'specmode','psd','logtrials','on'});
STUDY = pop_specparams(STUDY, 'averagechan','on', 'freqrange',[1 50]);
STUDY = std_specplot(STUDY,ALLEEG,'channels',{'Fp1','AF7','AF3','F1','F3','F5','F7','FT7','FC5','FC3','FC1','C1','C3','C5','T7','TP7','CP5','CP3','CP1','P1','P3','P5','P7','P9','PO7','PO3','O1','Iz','Oz','POz','Pz','CPz','Fpz','Fp2','AF8','AF4','AFz','Fz','F2','F4','F6','F8','FT8','FC6','FC4','FC2','FCz','Cz','C2','C4','C6','T8','TP8','CP6','CP4','CP2','P2','P4','P6','P8','P10','PO8','PO4','O2'}, 'design', 1);
- see code in std_spec and spectopo