in silico assembly of a TU vector
It is possible to assemble Yeast Pathway Kit vectors by hand using ApE or some other DNA editor. This require the user to carefully identify the relevant shared sequences between molecules and manipulate the sequences correctly. However it is slow, tedious and error prone.
An alternative is to use an automatic recombination simulator like the one in the pydna Python package.
For this exercise, we will use pydna and google colab which you can use if you have a free google account. Colab is a hosted Jupyter Notebook service that requires no setup. A Jupyter notebook is a python program file that can also show comments and images as well as intermediate results. Colab allows you to write and execute Python in your browser without installing any software.
The first step is to collect all sequences needed for the assembly. For a Yeast Pathway Kit single gene expression TU vector, this means:
- A linear plasmid sequence
- A promoter PCR product
- A gene PCR product
- A terminator PCR product
For this example, we will use the assembly of the expression vector pTA1_TDH3_ScATF1_PGI1.

This vector expresses the ATF1 gene from Saccharomyces cerevisiae using the TDH3 promoter and PGI1 terminator. The pTA1 vector provides selection markers and origin of replication.
The pTA1 vector is available here. It should be linearized using the ZraI restriction enzyme. The linear plasmid sequence can be obtained by transforming the sequence using the ApE Edit>"Linearize @ insert site" function.
The PCR products can be obtained using WebPCR and the PCR primers indicated in the table below. The primer sequences are available here.
| Target | Template | Forward primer | Reverse primer |
|---|---|---|---|
| Promoter | pYPKa_Z_TDH3 | 577 | 567 |
| Gene | [[pYPKa_A_ATF1 | in-silico-assembly-of-pYPKa_A_ATF1]] | 468 |
| Terminator | pYPKa_E_PGI1 | 568 | 578 |
Collect the linear vector sequence and the three PCR product sequences in sequence_formats#fasta format in a text editor such as Notepad like so:

Go to Google colab in you web browser. Create a new notebook by clicking on the "New notebook button", see the image below:

Copy the code below into the first cell. This code will tell the python package manager pip to install the pydna package which has the functionality we need.
!pip install pydna
Run the first cell by clicking the arrow button  and wait for the execution to finish (see below)).
and wait for the execution to finish (see below)).

You can ignore the output from this cell.
Click on the  button to get a new code cell (see below):
button to get a new code cell (see below):

Copy the code below into the new cell and execute.
from pydna import logo
from pydna.parsers import parse
from pydna.assembly import Assembly
logo()
You should now have a printout of the pydna logo:

Create a new code cell and paste the code below. Replace the sequences with your own and execute. Make sure that you adhere to the FASTA sequence format.
sequences = """\
>pTA1 linear
replace-this-text-with-your-own-DNA-sequence
>promoter
replace-this-text-with-your-own-DNA-sequence
>gene
replace-this-text-with-your-own-DNA-sequence
>terminator
replace-this-text-with-your-own-DNA-sequence
"""

There should be no output from the code cell above.
Create a new code cell and paste the code below and execute.
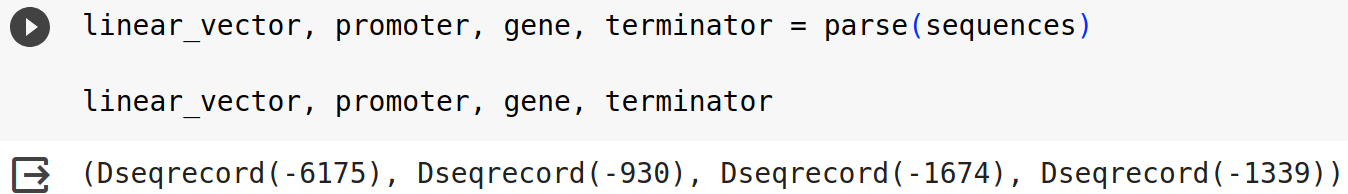
linear_vector, promoter, gene, terminator = parse(sequences)
linear_vector, promoter, gene, terminator
For this example, you should have an output like the one below.
asm = Assembly((linear_vector, promoter, gene, terminator))
candidates = asm.assemble_circular()
candidate, *rest = candidates
candidate.figure()
You should have a figure like this one as result:

Create a new code cell, paste the code below and execute.
result = candidate.synced("gttctgatcctcgagcatcttaagaattc")
result.name = "pTA1_TDH3_ScATF1_PGI1" # Change this name as needed
print(result.format())
This should give you a sequence of the plasmid in Genbank format:

Create a new code cell and paste the code below and execute.
result.cseguid()

Compare the cSEGUID code with the ones of your colleagues.