-
Notifications
You must be signed in to change notification settings - Fork 14
Description
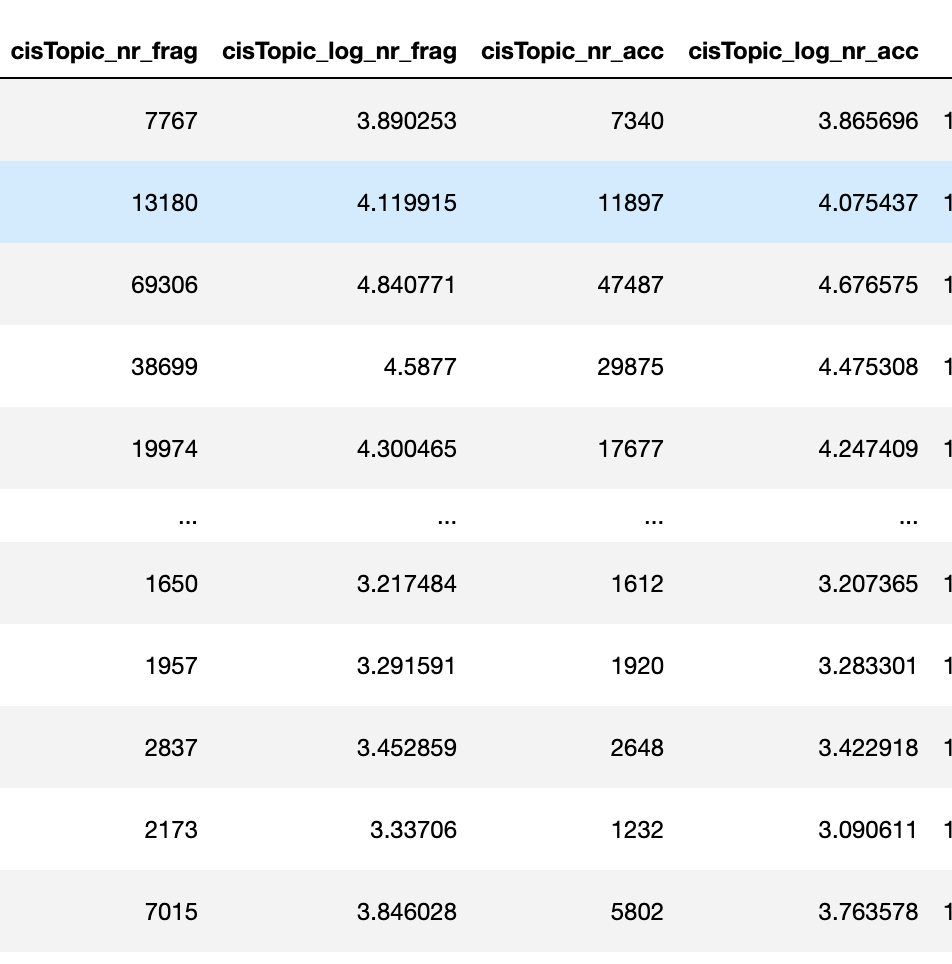
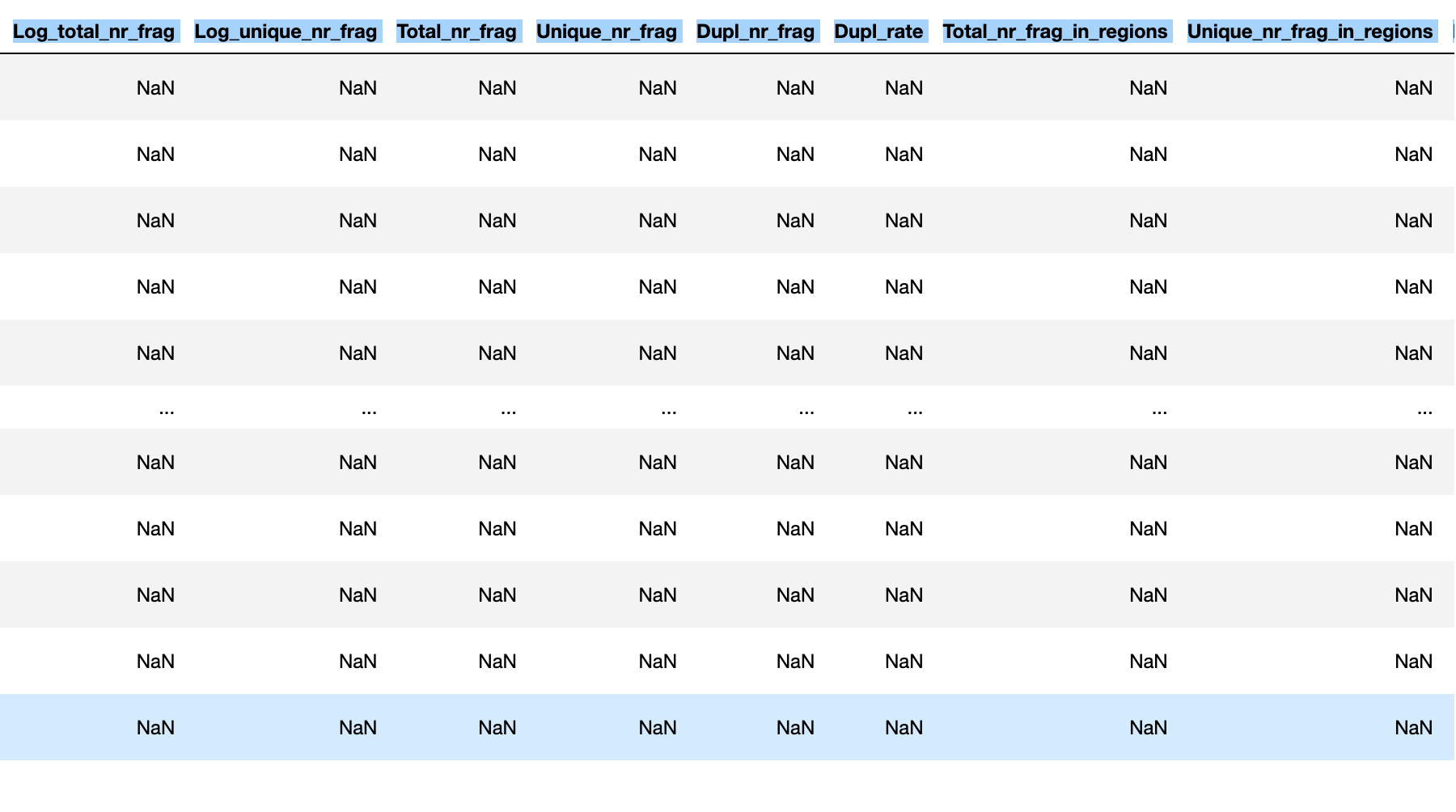
Hi there, I'm analyzing Dmel scATACseq for embrogenesis with pycistopic. Things went well until I checked cistopic object and all of the Log_total_nr_frag | Log_unique_nr_frag | Total_nr_frag | Unique_nr_frag | Dupl_nr_frag | Dupl_rate | Total_nr_frag_in_regions | Unique_nr_frag_in_regions | FRIP | TSS_enrichment were NaN for all rows.
I was wondering if there could possibly anything that I can work around this? Thank you,
Describe the bug
A clear and concise description of what the bug is.
To Reproduce
Commands relevant to reproduce the error.
Error output
Paste the entire output of the command, including log information prior to the error.
Expected behavior
A clear and concise description of what you expected to happen.
Screenshots
If applicable, add screenshots to help explain your problem or show the format of the input data for the command/s.
Version (please complete the following information):
- Python: [e.g. Python 3.7.4]
- If a bug is related to another module [e.g. matplotlib 3.3.9]
Additional context
Add any other context about the problem here.