-
Notifications
You must be signed in to change notification settings - Fork 7
Do continuous integration and codecoverage as in reduceSPI #19
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
…and Build not copy pasted, and DeepSource alredy present
…mpty in ioSPI and absent in reduceSPI
…version of pip (warning in continuous integration)
…ne should end with a period
…ll, but did not result in any comittable changes
|
I'm struggling to satisfy isort/flake8/black at the same time. what is the workflow? I'm isorting, but then flake8 has an issue |
|
Is this a possible fix? |
|
@geoffwoollard the pre-commit hook flake8 does not correct the files automatically, it is just failing. In this case, we need to write the error messages and address them. Here:
Does it work to just replace reduceSPI by ioSPI?
Let's use tests/test_*.py for now. We'll organize them in subfolders when we have more tests: bottom-up approach = we do it only when we need it.
Yay thanks.
Cool. thanks.
OK!
No, it's not meant to be like this: ioSPI/iotools is redundant, since we know that we will have iotools in ioSPI. Let's use the bottom up approach again, and only have one |
|
The problem is that flake8 wants And isort wants When I commit with the hooks, isort changes things to Then flake8 fails. These links say I need to tell flake8 somehow that there should be a line between them, because ioSPI is local, not third party. |
…tatement, otherwise tries to put together. But isort knows it is local so automatically edits
|
Actually this works |
.flake8
Outdated
| docstring-convention = numpy | ||
| import_order_style = smarkets | ||
| max-line-length = 88 | ||
| extend-ignore = E203 |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@geoffwoollard how about adding I202 to the extend-ignore line?
…ts own __init__.py file and it can not go above that level to ioSPI
…are in the wrong order. 'import ioSPI.iotools.cryo_dataset' should be before 'import torch'
…hange back name after pass test
|
I was really confused for a while that the CI couldn't find this module. Locally it was in ioSPI, but actually git saw it as in iospi. So I just copied it (cp, not git mv) and renamed it as cryodataset.py. 2b78842 |
|
@fredericpoitevin maybe you can write some tests for |
Codecov Report
@@ Coverage Diff @@
## master #19 +/- ##
=========================================
Coverage ? 66.32%
=========================================
Files ? 4
Lines ? 190
Branches ? 0
=========================================
Hits ? 126
Misses ? 64
Partials ? 0 Continue to review full report at Codecov.
|
 ninamiolane
left a comment
ninamiolane
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Looking good! We can merge this since most of it is working, and it will give us a chance to see if everything else is in place at the next PR.
But only if you can tackle the 90% code coverage threshold ASAP?
.codecov.yml
Outdated
| project: | ||
| default: | ||
| target: 90% | ||
| target: 50% |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This should be 90% though: this is the threshold under which the PR will not pass: we don't want a PR to pass if we don't have a least 90% of the code that is covered, i.e. tested by the unit-tests. Why the decrease here?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Ok I'll see if I can write some tests for Fred's atomic_models.py
.codecov.yml
Outdated
| default: | ||
| # basic | ||
| target: 90% | ||
| target: 50% |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Ditto
| ignore: | ||
| - "data" | ||
| - "coverage" | ||
| - "notebooks" |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
We will have to test the notebooks actually, but we can leave this for later I guess.
| docstring-convention = numpy | ||
| import_order_style = smarkets | ||
| max-line-length = 88 | ||
| extend-ignore = E203,I202,I100 |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Just curious, what is I100? Do you know why I didn't need these exclude in reduceSPI?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
It's very similar as I202, with the spacing between the imports. Flake does not know that ioSPI is local, and wants to put the imports together, while isort knows and splits them up. See 2d7e03c
In your tests you do https://github.com/compSPI/reduceSPI/blob/master/tests/test_offline.py#L3, but there is no other import like import numpy or something nearby. My guess is that you would have the same issue.
|
I wrote some tests, but the CI is not happening? Where did they go? |
|
Oh yeah, I had the same problem with reduceSPI at some point... and then it came back! I don't know what is happening, TBH it could be that github action is experiencing a bug, or latency? Maybe let's try to merge this, and see what it gives on a new PR? |
|
Oh lol, it's back x) |
|
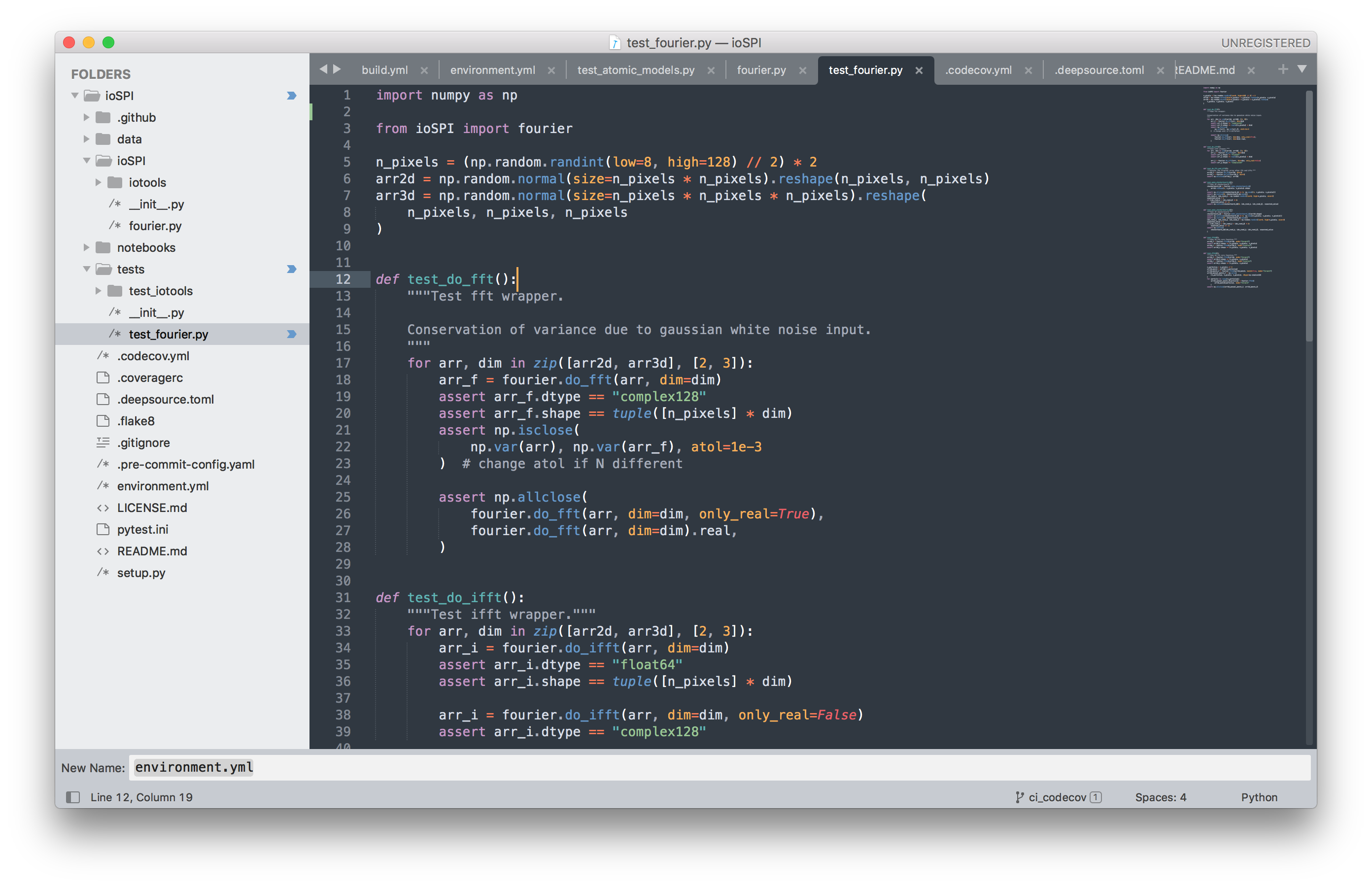
There's some problem with the code coverage. I wrote tests for parts that is says aren't covered. They are special because they are @Jitted, so maybe there's some issue with that. The tests are https://github.com/geoffwoollard/ioSPI/blob/ci_codecov/tests/test_fourier.py#L51 and https://github.com/geoffwoollard/ioSPI/blob/ci_codecov/tests/test_fourier.py#L65 |
|
Let's merge and see what codecov says. Thanks for this 🎉 |
Some notes on what I did that deserve a closer look
Badges in README.md. What am I supposed to use for the links at the top of https://raw.githubusercontent.com/compSPI/reduceSPI/master/README.md
reduceSPI just has tests in
tests/test_*.py. However ioSPI hastests/test_*.pyand alsotests/test_iotools/test_*.py. What should be inioSPI/ci_codecov/pytest.inito handle the extra folder? geoffwoollard@b668095I took the isort and black transformers out of
.deepsource.tomlgeoffwoollard@d99cc15.codecov.ymlhas an extra part to ignore folders that are absent in reduceSPI. I removedcoveragefolder because its empty in ioSPI and absent in reduceSPI. geoffwoollard@1980c1e.github/workflows/build.ymlis quite a bit different. Note that the flags for black are absent. geoffwoollard@6311a51#diff-5c3fa597431eda03ac3339ae6bf7f05e1a50d6fc7333679ec38e21b337cb6721L51 These were important to have the linting and testing pass.Aside: there's a folder
iospi/iotoolsand alsoioSPI/iotools/. They have different .py files in them. Is it supposed to be like that?