-
Couldn't load subscription status.
- Fork 674
Description
Dear There,
I recently re-installed the single cell environment. Then, the example code of sc.tl.marker_gene_overlap() on scanpy tutorial does not work. It give the same values to each cluster. Thanks for your help!
Lianyun
import scanpy as sc
adata = sc.datasets.pbmc68k_reduced()
sc.pp.pca(adata, svd_solver='arpack')
sc.pp.neighbors(adata)
sc.tl.louvain(adata)
sc.tl.rank_genes_groups(adata, groupby='louvain')
marker_genes = {'CD4 T cells': {'IL7R'},
'CD14+ Monocytes': {'CD14', 'LYZ'},
'B cells': {'MS4A1'},
'CD8 T cells': {'CD8A'},
'NK cells': {'GNLY', 'NKG7'},
'FCGR3A+ Monocytes': {'FCGR3A', 'MS4A7'},
'Dendritic Cells': {'FCER1A', 'CST3'},
'Megakaryocytes': {'PPBP'}}
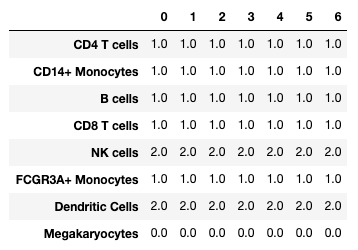
marker_matches = sc.tl.marker_gene_overlap(adata, marker_genes) 0 1 2 3 4 5 6
CD4 T cells 1.0 1.0 1.0 1.0 1.0 1.0 1.0
CD14+ Monocytes 1.0 1.0 1.0 1.0 1.0 1.0 1.0
B cells 1.0 1.0 1.0 1.0 1.0 1.0 1.0
CD8 T cells 1.0 1.0 1.0 1.0 1.0 1.0 1.0
NK cells 2.0 2.0 2.0 2.0 2.0 2.0 2.0
FCGR3A+ Monocytes 1.0 1.0 1.0 1.0 1.0 1.0 1.0
Dendritic Cells 2.0 2.0 2.0 2.0 2.0 2.0 2.0
Megakaryocytes 0.0 0.0 0.0 0.0 0.0 0.0 0.0
Versions
anndata 0.7.4
scanpy 1.6.0
sinfo 0.3.1
PIL 7.2.0
anndata 0.7.4
backcall 0.2.0
cffi 1.14.1
cycler 0.10.0
cython_runtime NA
dateutil 2.8.1
decorator 4.4.2
get_version 2.1
h5py 2.10.0
igraph 0.8.2
ipykernel 5.3.4
ipython_genutils 0.2.0
jedi 0.15.2
joblib 0.16.0
kiwisolver 1.2.0
legacy_api_wrap 1.2
llvmlite 0.34.0
louvain 0.7.0
matplotlib 3.3.1
mpl_toolkits NA
natsort 7.0.1
numba 0.51.2
numexpr 2.7.1
numpy 1.19.1
packaging 20.4
pandas 1.1.1
parso 0.5.2
pexpect 4.8.0
pickleshare 0.7.5
pkg_resources NA
prompt_toolkit 3.0.7
ptyprocess 0.6.0
pycparser 2.20
pygments 2.6.1
pyparsing 2.4.7
pytz 2020.1
scanpy 1.6.0
scipy 1.5.2
setuptools_scm NA
sinfo 0.3.1
six 1.15.0
sklearn 0.23.2
statsmodels 0.12.0
storemagic NA
tables 3.6.1
texttable 1.6.3
tornado 6.0.4
traitlets 4.3.3
umap 0.4.6
wcwidth 0.2.5
zmq 19.0.2
IPython 7.18.1
jupyter_client 6.1.7
jupyter_core 4.6.3
notebook 6.1.3
Python 3.8.5 | packaged by conda-forge | (default, Aug 29 2020, 01:22:49) [GCC 7.5.0]
Linux-3.10.0-1062.9.1.el7.x86_64-x86_64-with-glibc2.10
128 logical CPU cores, x86_64
Session information updated at 2020-09-09 15:47
Metadata
Metadata
Assignees
Labels
No labels