-
Notifications
You must be signed in to change notification settings - Fork 25
Description
Greetings,
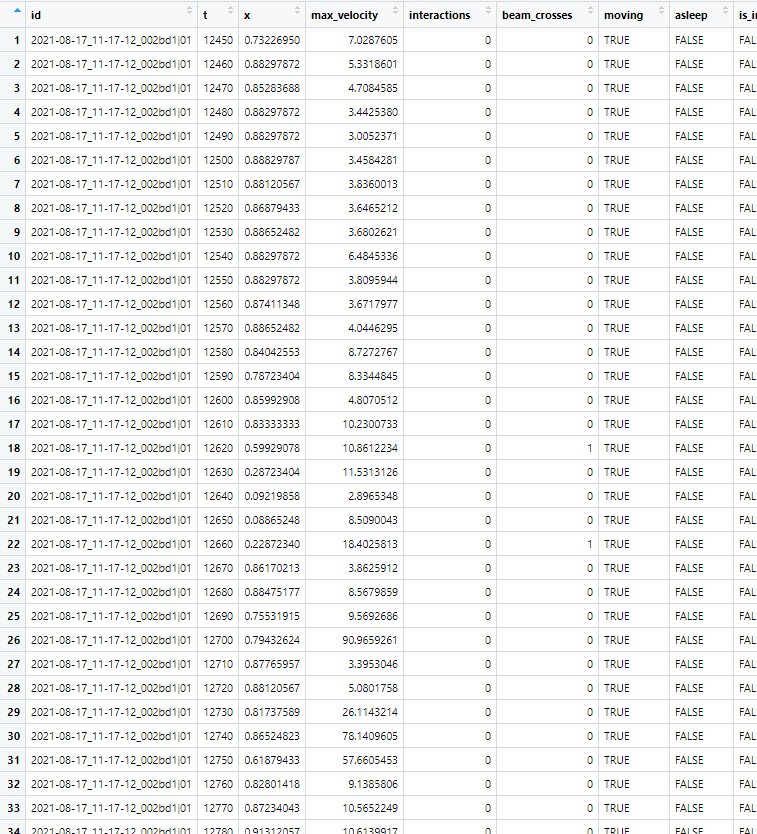
Me and my lab mates have been using the Ethoscope to study sleep in several Drosophila mutants. As it can be expected, the sleep data we analyzed based on the DAM system is different from that of the Ethoscope. Thus, we wanted to analyze sleep in Ethocope in DAM fashion to better understand the difference. Hence, we wanted to analyze sleep not in terms of not-moving for 5 mins (300 sec), but in terms of not-crossing the beam for 5 min. As it can be seen in the raw Ethoscope data (image attached), beam_crosses information is provided in 10 sec interval alongside other parameters. Since I am new to R, I don't know efficient ways to make this analysis. However, I just tried the following double loop to compute a data table for sleep that is based on beam crosses:
initialize the data table; dt1 is my raw data from the Ethoscope;
sleep_check <- data.table((dt1[1,"id"]), (dt1[1, "t"]), asleep=paste("FALSE"))
##since the first row is initialized start the loop from second row
for (row in 2:nrow(dt1)){
id_name=dt1[row, "id"]
time=dt1[row, "t"]
count=row
dt_tempo<- data.table(id_name, time, asleep="TRUE")
while (count<row+30 && count<nrow(dt1)){
##loop for 30 data points to analyze beam_crosses for 300 second
beam_crossed=dt1[count,"beam_crosses"]
count=count+1
if (beam_crossed>0)
{
dt_tempo<- data.table(id_name, time, asleep=paste("FALSE"))
break
}
}
sleep_check <- rbind(sleep_check, dt_tempo)
}
However, since my raw data has more than 3,000,000 rows, the computation is running for a very long time, like more than 20 hours. I wanted to ask if you can show me a better way of approaching this problem.
Thanks very much