-
Notifications
You must be signed in to change notification settings - Fork 687
Closed
Labels
Description
- I have checked that this issue has not already been reported.
- I have confirmed this bug exists on the latest version of scanpy.
- (optional) I have confirmed this bug exists on the master branch of scanpy.
When running the Integrating spatial data with scRNA-seq using scanorama tutorial with the provided sample data, the following error occurs.
This may be related to the following warning I also see
<string>:6: VisibleDeprecationWarning: Creating an ndarray from nested sequences exceeding the maximum number of dimensions of 32 is deprecated. If you mean to do this, you must specify 'dtype=object' when creating the ndarray.
How can I overcome this issue?
Example code below has how I downloaded the data in the Data integration and label transfer from scRNA-seq dataset section of the tutorial and then just the code block where the error actually occurs.
Minimal code sample (that we can copy&paste without having any data)
import subprocess
from pathlib import Path
if Path('./downloaded_data/adata_processed.h5ad').is_file():
print("Data previously downloaded, skipping to next step")
else:
subprocess.run(['wget', '-O', './downloaded_data/adata_processed.h5ad', 'https://hmgubox.helmholtz-muenchen.de/f/4ef254675e2a41f89835/?dl=1'])
adata_cortex = sc.read("./downloaded_data/adata_processed.h5ad")
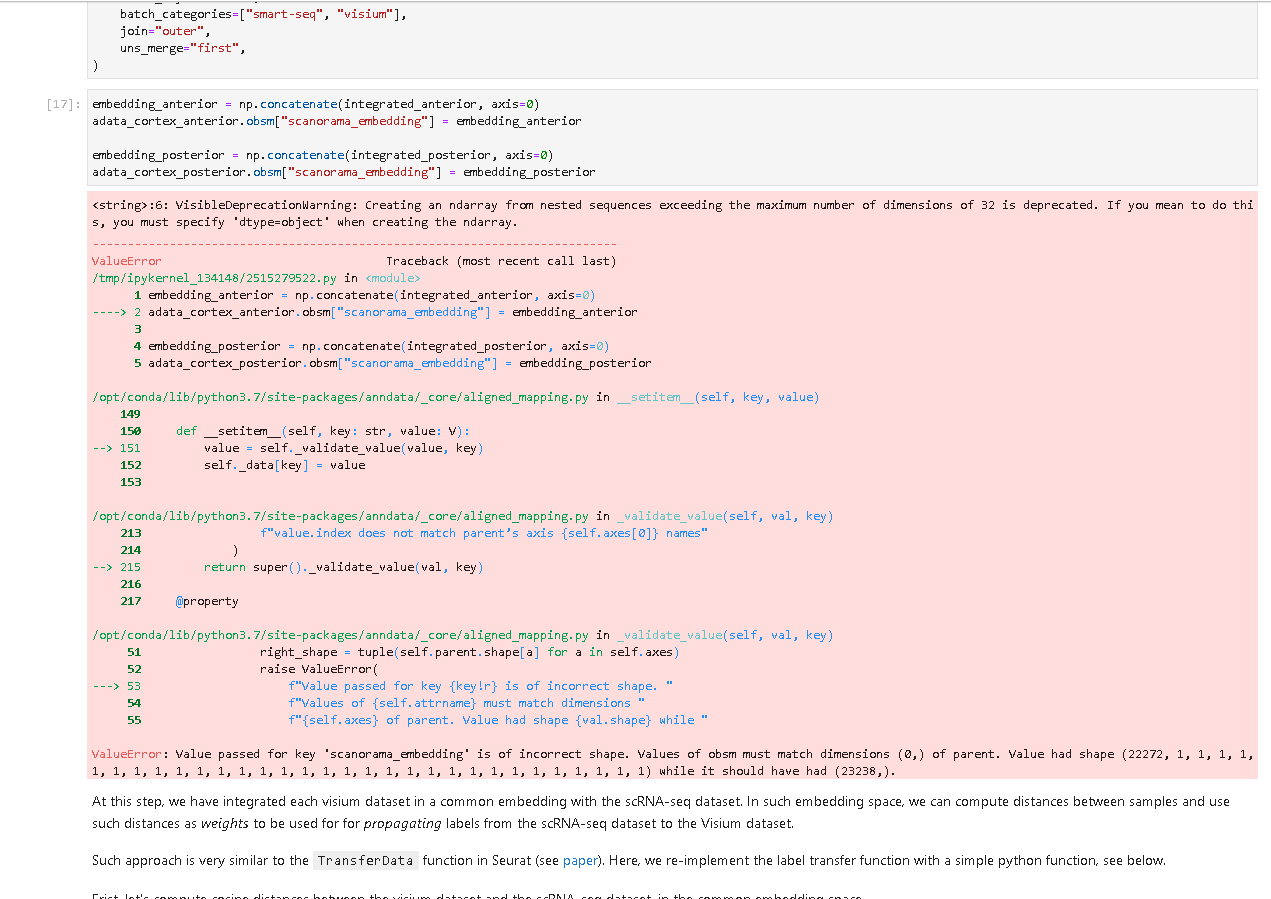
embedding_anterior = np.concatenate(integrated_anterior, axis=0)
adata_cortex_anterior.obsm["scanorama_embedding"] = embedding_anterior
embedding_posterior = np.concatenate(integrated_posterior, axis=0)
adata_cortex_posterior.obsm["scanorama_embedding"] = embedding_posterior<string>:6: VisibleDeprecationWarning: Creating an ndarray from nested sequences exceeding the maximum number of dimensions of 32 is deprecated. If you mean to do this, you must specify 'dtype=object' when creating the ndarray.
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
/tmp/ipykernel_134148/2515279522.py in <module>
1 embedding_anterior = np.concatenate(integrated_anterior, axis=0)
----> 2 adata_cortex_anterior.obsm["scanorama_embedding"] = embedding_anterior
3
4 embedding_posterior = np.concatenate(integrated_posterior, axis=0)
5 adata_cortex_posterior.obsm["scanorama_embedding"] = embedding_posterior
/opt/conda/lib/python3.7/site-packages/anndata/_core/aligned_mapping.py in __setitem__(self, key, value)
149
150 def __setitem__(self, key: str, value: V):
--> 151 value = self._validate_value(value, key)
152 self._data[key] = value
153
/opt/conda/lib/python3.7/site-packages/anndata/_core/aligned_mapping.py in _validate_value(self, val, key)
213 f"value.index does not match parent’s axis {self.axes[0]} names"
214 )
--> 215 return super()._validate_value(val, key)
216
217 @property
/opt/conda/lib/python3.7/site-packages/anndata/_core/aligned_mapping.py in _validate_value(self, val, key)
51 right_shape = tuple(self.parent.shape[a] for a in self.axes)
52 raise ValueError(
---> 53 f"Value passed for key {key!r} is of incorrect shape. "
54 f"Values of {self.attrname} must match dimensions "
55 f"{self.axes} of parent. Value had shape {val.shape} while "
ValueError: Value passed for key 'scanorama_embedding' is of incorrect shape. Values of obsm must match dimensions (0,) of parent. Value had shape (22272, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1) while it should have had (23238,).