-
Notifications
You must be signed in to change notification settings - Fork 429
Description
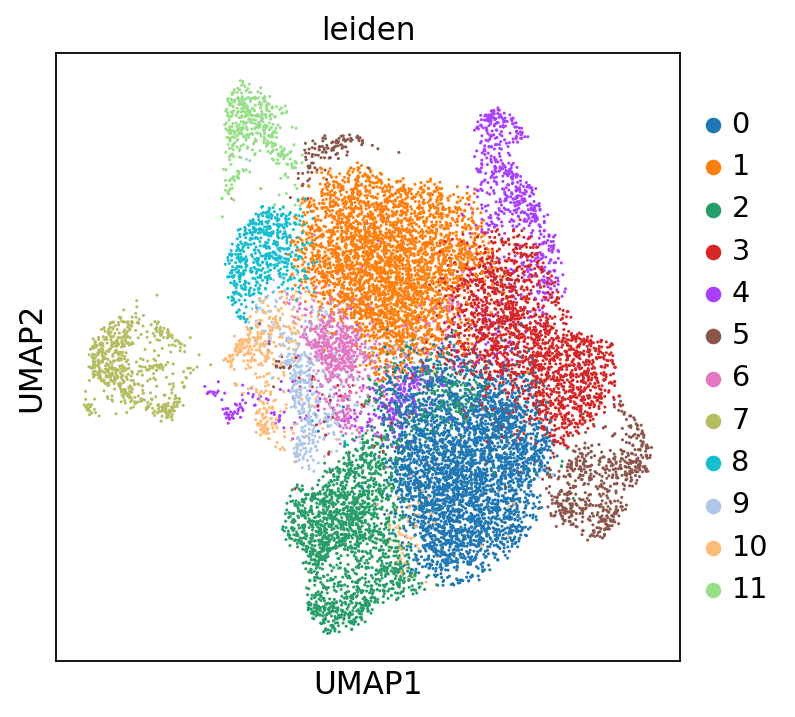
I trained an scVI model on a machine with an AMD EPYC 7352 CPU and a Quadro RTX 8000 GPU and saved the model with model.save(). When I load the model on the same machine and retrieve the latent representation, the resulting UMAP is different to when I load the model on a different machine with a Intel(R) Xeon(R) CPU E7-4850 v4 CPU and no GPU.
Is that expected? I was hoping to share the trained model alongside my code to make the analysis reproducible. If so, what's the recommended way to share reproducible analyses with scVI? (I am aware that training the model is not expected to be reproducible).
Here's the code I use to retrieve the latent representation and compute the UMAP:
model = scvi.model.SCVI.load(f"{output_dir}/scvi_model_all_genes", adata_scvi)
adata.obsm["X_scVI"] = model.get_latent_representation(adata_scvi)
sc.pp.neighbors(adata, use_rep="X_scVI", n_neighbors=15)
sc.tl.umap(adata)Versions:
0.8.1
Model loaded on the AMD machine:
Model loaded on the Intel machine:
(Neighbors and UMAP were computed on the intel machine in both cases to make sure the instability is not in neighbors and umap)