-
Notifications
You must be signed in to change notification settings - Fork 131
Automatic documentation for experiment and analysis #116
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Automatic documentation for experiment and analysis #116
Conversation
This reverts commit eb60945.
 eggerdj
left a comment
eggerdj
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is some neat functionality. I did not review the RB experiments but focused on Spec.
| from . import analysis_docs, experiment_docs, set_option_docs | ||
| from .descriptions import OptionsField, Reference, CurveFitParameter, to_options |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Most of the package seems to use absolute imports, why relative here'
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@chriseclectic suggested to use relative import in another PR. I prefer absolute one but we don't have standard.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I also prefer absolute
| return writer.docstring | ||
|
|
||
|
|
||
| class CurveAnalysisDocstring(_DocstringMaker): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Can this somehow inherite from StandardAnalysisDocstring? This would avoid some code duplication. E.g.
if warning:
writer.write_warning(warning)
appears in both.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I understand your point, but we cannot turn this into a subclass. Because order of calling writer method is important to standardize docstring section ordering. Although we can make standard one a superclass of this, we need to write full make_docstring method for this and there is almost no benefit from doing so.
| @abstractmethod | ||
| def make_docstring(cls, *args, **kwargs) -> str: | ||
| """Write a docstring.""" | ||
| pass |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Do we need pass here?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
No really. But I see some other abstract methods have pass (maybe personal preference).
Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com>
Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com>
Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com>
|
Yes, thanks for the feedback @wshanks ! I agree IDE experience should be improved in feature. We can take two approaches AFAIK
I think we need to continue the discussion. I'll write an issue to track this issue. |
|
The sphinx build error was due to missing documentation (and error) in With above fix, we can generate html documentation for RB as shown in here. This is just MVP and you need to properly update documentation with custom section and directives like this. I think we can do this in follow-up PR. Let's do dockathon! :) |
 eliarbel
left a comment
eliarbel
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
From usage perspective this looks good.
It would be good to allow seamless transition between existing docstring to the auto docstring generation format. To this end I recommend:
- Use the same indentation rules, i.e. don't force un-indenting actual text (e.g. the text under Overview)
- Use the same new line rules, i.e. don't force adding new lines between the
# section: <>lines and the actual text
- add template for example - section indent and rf rule update
…ure/auto-option-docs-gen
|
@eliarbel Thank you so much for usability testing. Happy to hear your positive feedback.
Sure. Now we can use indented text block. The parser can still understand where is the section document even if indent is missing. This allows you to write document section in the exactly the same format as before.
Actually it doesn't force adding the line feed (in contrast to Sphinx that forces us to remove empty line after section statement -- without error message...). You can see updated example package. I just removed empty line (now it really looks like previous format) and it still works. These changes are made in this commit f6cda8f You can also check updated RB example here -- please ignore two files in the beginning. |
- add extra indent check - autofix section indent error
…ure/auto-option-docs-gen
 eliarbel
left a comment
eliarbel
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think this is ready for production!
|
@eggerdj this is pending your approval, can you give a look? |
* ResultsDB tutorial (#208) * added warning and test * added tutorial * formatting * added load data * changed to experimentdata * updated loade data * changed load syntax * Calibrations tutorial (#184) * * Added calibrations tutorial. * Added time-zone fix in Calibrations. * * Propagating timezone change. * * Docs for NB. * * NB rewording. * * Test math. * * Math in DRAG. * Update docs/tutorials/rb_example.ipynb * Update docs/tutorials/qst_example.ipynb * Update docs/tutorials/qst_example.ipynb * Update docs/tutorials/qst_example.ipynb * Update docs/tutorials/qv_example.ipynb * * Added comment on parameters and channels. * Update docs/tutorials/calibrating_armonk.ipynb Co-authored-by: Will Shanks <wshaos@posteo.net> * Update docs/tutorials/calibrating_armonk.ipynb Co-authored-by: Will Shanks <wshaos@posteo.net> * * Added comment on Amplitude.update. * * DRAG text. * * Added comment on qubit frequencies. * * Added change and comment to show the initial guess values. * * Updated Armonk NB with xm schedule and unlinked amp and beta for xp and x90p. Co-authored-by: Will Shanks <wshaos@posteo.net> Co-authored-by: Helena Zhang <Helena.Zhang@ibm.com> * Fix errors in QST notebook (#214) * Make `block_for_results` return self * Fix errors in qst notebook so it runs Co-authored-by: Helena Zhang <Helena.Zhang@ibm.com> * Automatic documentation for experiment and analysis (#116) * initial commit model based experiment * Revert "initial commit model based experiment" This reverts commit eb60945. * wip options field * fix bug * update type handling * black * add experiment options update and class docstring update * cleanup facades and make autodoc module a package * wip analysis documentation * update analysis docs and reformat styles. * black and typing * wip lint * replace LF * strip LFs of input text * update irb docstring * ignore lint * unittest * Update qiskit_experiments/analysis/curve_analysis.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Sphinx autodoc extension for documentation * revert experiment modules * remove old autodoc module * fix old import * remove change to base analysis * update docs * lint * add example of analysis default options * move package under docs * fix import path * fix missing docs * comment from Eli - add template for example - section indent and rf rule update * fix indent correction logic * update example * robust to indent error - add extra indent check - autofix section indent error * fix import paths * fix example documentation Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> Co-authored-by: Helena Zhang <Helena.Zhang@ibm.com> * Remove bounds from T1 (#168) * Remove bounds from T1 * reverting the tutorial Co-authored-by: Helena Zhang <Helena.Zhang@ibm.com> * Set minimum python version to 3.7 (#227) This commit changes the minimum supported python version from 3.6 to 3.7. qiskit-experiments uses dataclasses internally which was a python feature introduced in 3.7 and the code as packaged doesn't actually work on 3.6. This hasn't been caught in CI because a test dependency of qiskit-experiments is installing the dataclasses backport package as a dependency which is masking this issue. While we can list that package as a depedency of qiskit-experiments too to resolve this, python 3.6 is deprecated in the upstream qiskit packages and the next qiskit-terra minor version release (0.19.0) will be the last to support it. At this point it just makes more sense to drop 3.6 support. At the same time the minimum supported version of the qiskit-terra package is bumped to the latest release 0.18.0 which is actually the minimum version needed (we were installing from git in CI to workaround this pre-0.18.0 release) and 0.17.0 was the release which deprecated python 3.6 in qiskit which lines up well with dropping 3.6 support here. * Fix headers in RB tutorial (#223) Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> Co-authored-by: Will Shanks <wshaos@posteo.net> Co-authored-by: Christopher J. Wood <cjwood@us.ibm.com> Co-authored-by: Naoki Kanazawa <knzwnao@jp.ibm.com> Co-authored-by: Yael Ben-Haim <yaelbh@il.ibm.com> Co-authored-by: Matthew Treinish <mtreinish@kortar.org> Co-authored-by: Shelly Garion <46566946+ShellyGarion@users.noreply.github.com>
…#116) * initial commit model based experiment * Revert "initial commit model based experiment" This reverts commit eb60945. * wip options field * fix bug * update type handling * black * add experiment options update and class docstring update * cleanup facades and make autodoc module a package * wip analysis documentation * update analysis docs and reformat styles. * black and typing * wip lint * replace LF * strip LFs of input text * update irb docstring * ignore lint * unittest * Update qiskit_experiments/analysis/curve_analysis.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Update qiskit_experiments/characterization/qubit_spectroscopy.py Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> * Sphinx autodoc extension for documentation * revert experiment modules * remove old autodoc module * fix old import * remove change to base analysis * update docs * lint * add example of analysis default options * move package under docs * fix import path * fix missing docs * comment from Eli - add template for example - section indent and rf rule update * fix indent correction logic * update example * robust to indent error - add extra indent check - autofix section indent error * fix import paths * fix example documentation Co-authored-by: Daniel Egger <38065505+eggerdj@users.noreply.github.com> Co-authored-by: Helena Zhang <Helena.Zhang@ibm.com>
Summary
This PR adds auto docstring generation decorator for experiment and analysis classes.
close #92
Details and comments
The new package
qiskit_experiments.autodocsis added.Class implementation has changed. Look this comment.
#116 (comment)
-- old --
The docstring generator is implemented based on facade pattern which is often used for auto generation of, for example, webpage. This includes actual docstring writer class and facade class to define the style of docstring. By adding new facade class, you can define own docstring format for your subset of experiments.
Now base classes have several class attributes
__doc_*__corresponding to a section of documentation. A developer needs to fill these attributes to generate docstring sections. The allocation of sections are uniquely determined by the facade class, thus we can remove variation of documentation quality by developers.The actual usage of new attributes is shown in the
test.test_autodocs.pyalong with the unittest (This is good place to start review).-- old --
Alternative approaches
Option1: Copy-paste all option descriptions from the base class.
Option2: Just add reference to base class, i.e. See :class:`qiskit_experiment.MyBaseClass` for other options.
**fieldsand no option name is listed there. Thus, this approach drastically hurts usability.Considering cons of 1&2, both approaches are not realistic, in terms of our bandwidth for code maintenance and usability.
Examples
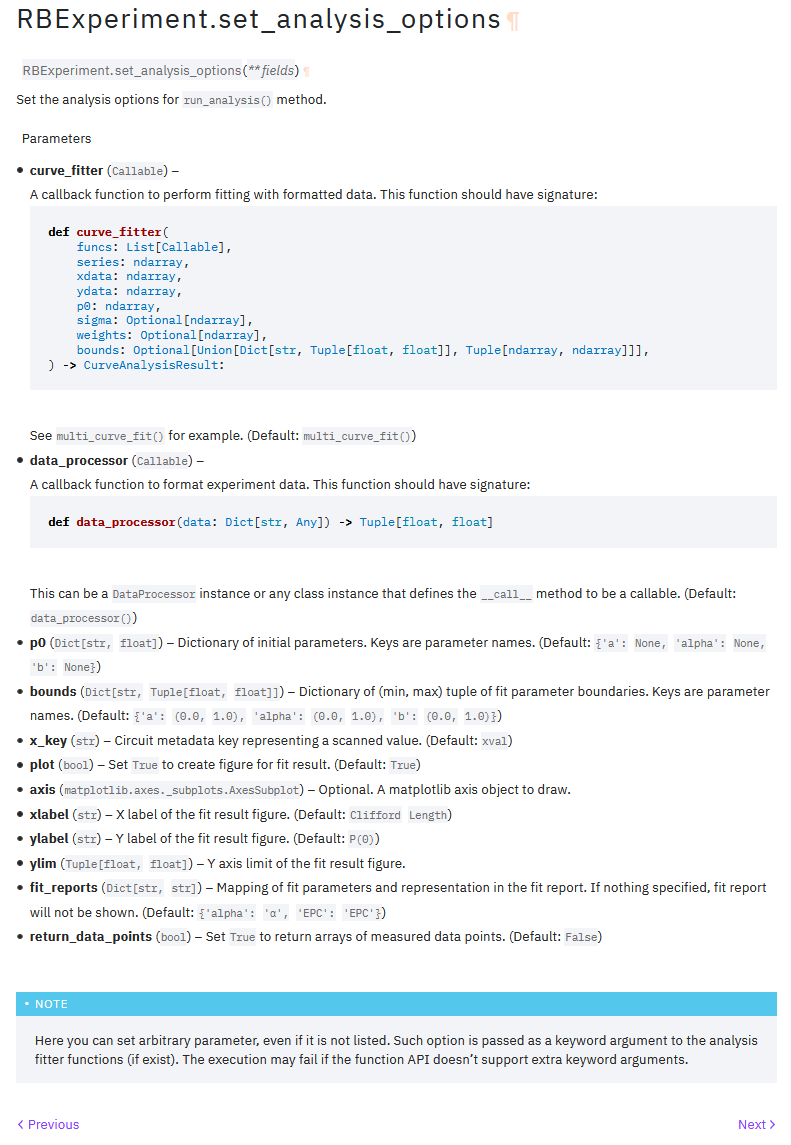
This is an example of automatically generated docstring. Seems like acceptable quality.
RB Experiment documentation
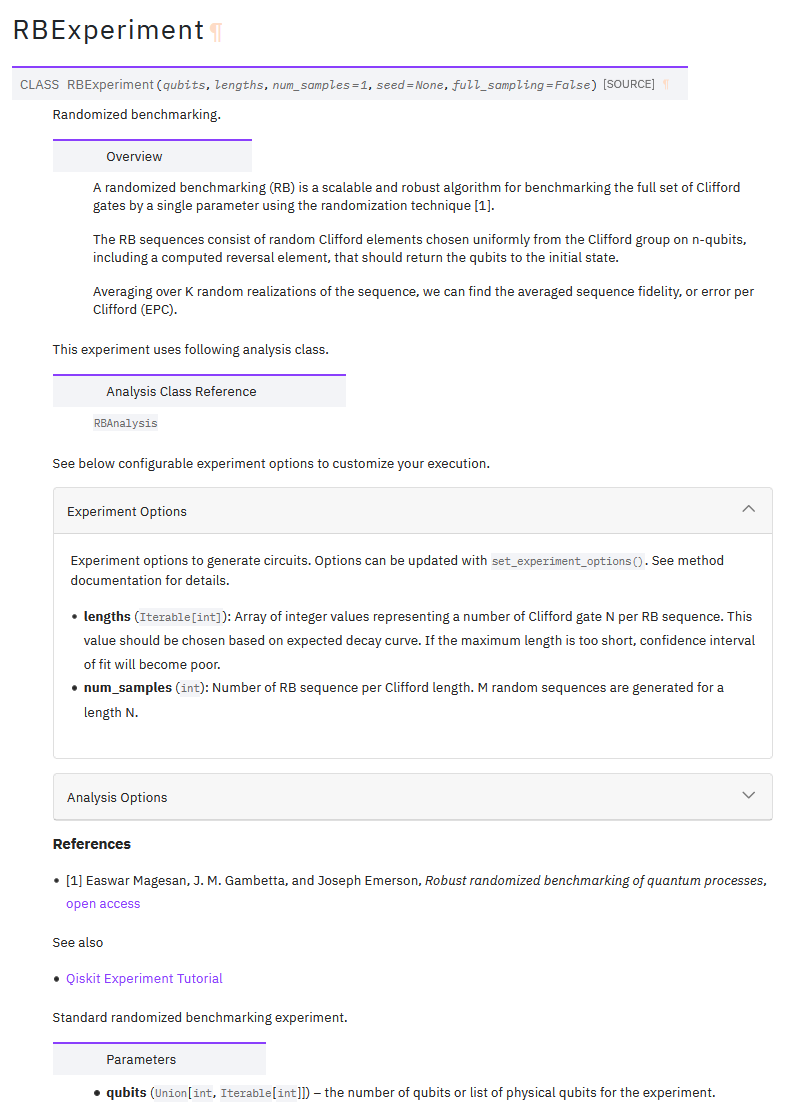
Set experiment options method

Set analysis options method
Analysis documentation
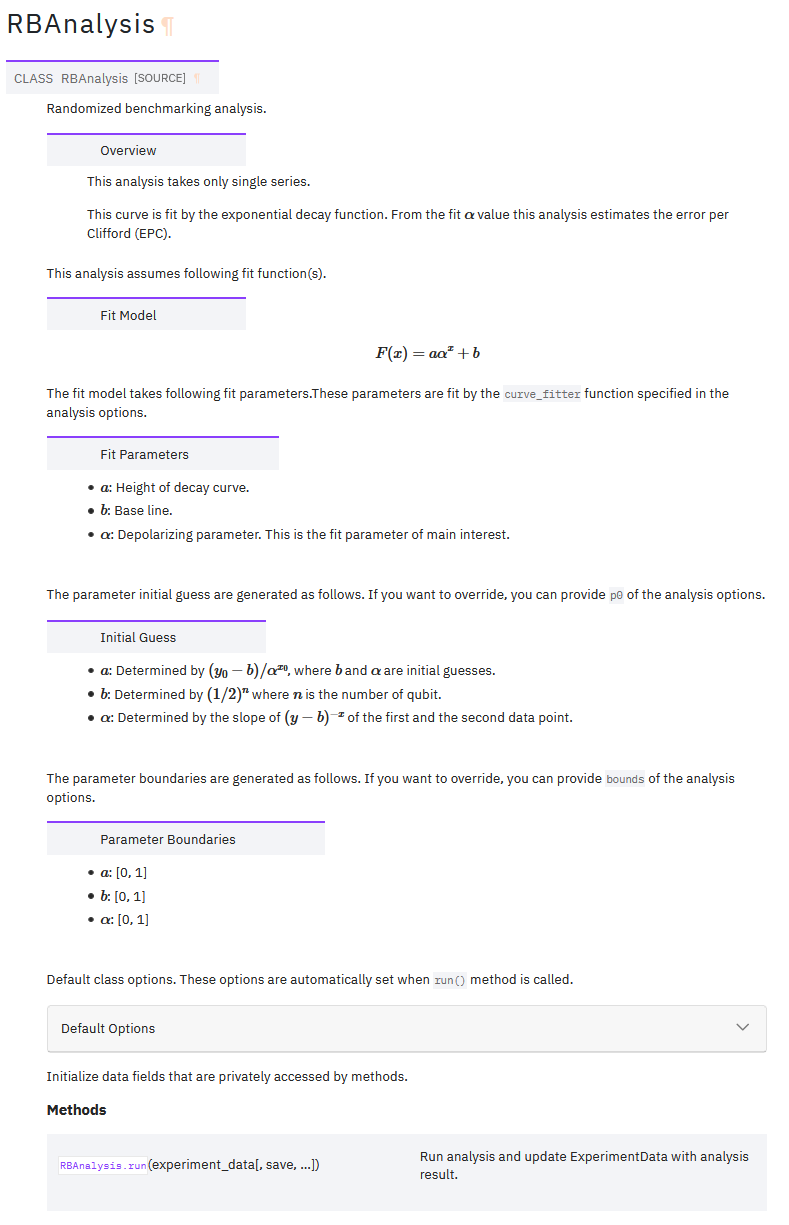
These are automatically generated documentations.